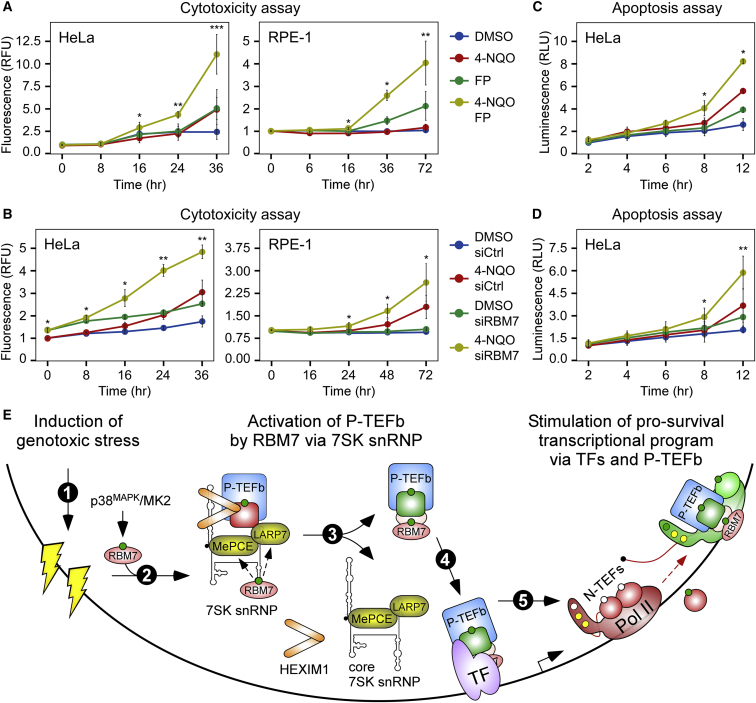
Figure 7.
P-TEFb and RBM7 Promote Cell Viability upon Genotoxic Stress
(A and B) Hypersensitivity of HeLa and RPE-1 cells to 4-NQO upon FP treatment (A) and RBM7 depletion (B). The cells were treated as indicated by the legends and examined at the time points indicated below the graphs. Two independent siRNAs (siRBM7 #2, HeLa cells; siRBM7 #1, RPE-1 cells) were used to deplete RBM7. Cytotoxicity results are presented as fluorescence values relative to the untreated control and plotted as the mean ± SEM (n = 3). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001, determined by Student’s t test using 4-NQO and 4-NQO FP or 4-NQO siRBM7 datasets, respectively.
(C and D) FP treatment (C) and RBM7 depletion (D) enhance 4-NQO-induced apoptosis in HeLa cells. The cells were treated as indicated by the legends and examined at the time points indicated below the graphs. siRBM7 #2 was used to deplete RBM7. Apoptosis results are presented as luminescence values relative to the untreated control and plotted as the mean ± SEM (n = 3). ∗p < 0.05; ∗∗p < 0.01, determined by Student’s t test using 4-NQO and 4-NQO FP or 4-NQO siRBM7 datasets, respectively.
(E) Model of P-TEFb activation by RBM7 during DDR. Genotoxic stress (step 1) provokes phosphorylation (green circle) of RBM7 by the p38MAPK-MK2 pathway. Subsequently, this triggers enhanced interaction of RBM7 with the core of 7SK snRNP (step 2; dashed arrows indicate interactions of RBM7 with MePCE and LARP7), triggering the release of inactive P-TEFb (CDK9 in red) from the core (step 3), yielding active P-TEFb (CDK9 in green). In turn, transcription factors (TFs) capture P-TEFb on chromatin (step 4). Stimulation of pro-survival DDR gene transcription at the Pol II pause release phase ensues (step 5), which is achieved by P-TEFb-mediated phosphorylation (green circles) of Pol II CTD at Ser2 as well as the negative transcription elongation factors (N-TEFs) NELF and DSIF. While NELF dissociates from Pol II, DSIF becomes a positive transcription elongation factor.