Abstract
Background
Hereditary spastic paraplegias (HSP) are of great clinical and genetic heterogeneity. According to the clinical features, HSP can be divided into pure or complicated subtypes which combined with other neurological symptoms including cerebellar ataxia. Up to date, 78 loci or genes have been implicated in HSP. CAPN1 was a novel gene detected recently for spastic paraplegia 76 (SPG76).
Methods
Patients referred to our clinic with spastic or spastic-ataxic gait were collected. Genetic testing of the probands were performed by target sequencing of a panel containing over 4000 known virulence genes. And the candidate mutations were further confirmed by polymerase chain reaction (PCR) and Sanger sequencing. The clinical materials of these patients were demonstrated retrospectively.
Results
Two Chinese patients, both from consanguineous families, each carried a novel homozygous mutation of CAPN1, p.R48X and p.R339X. The male proband presented pure HSP subtype while the female proband presented complicated HSP symptoms with cerebellar ataxia. We then reviewed all the literatures of HSP patients carrying CAPN1 mutations and summarized the molecular spectrum and clinical characteristics of CAPN1-related SPG76.
Conclusion
These two SPG76 patients carrying CAPN1 mutations were the first reported in China. By reviewing the clinical manifestations of SPG76 patients, we validated the “spastic-ataxia” phenotype and emphasized the association between spasticity and ataxia, indicating the importance of CAPN1 screening in HSP patients.
Keywords: Hereditary spastic paraplegias (HSP), Spastic paraplegia 76(SPG76), CAPN1 mutations, Ataxia
Introduction
Hereditary spastic paraplegias (HSP) present great genetic and clinical heterogeneity, mainly manifesting as spasticity and weakness in the lower limbs [1]. On the basis of clinical features, HSP can be categorized into pure and complicated subtypes [2]. In addition to the dominant progressive spasticity and weakness, pure HSP can also present symptoms of hypertonic bladder and sensory disturbances. Complicated HSP is often accompanied by other neurological symptoms, including cerebellar ataxia, seizure, extrapyramidal signs, intellectual disability, peripheral neuropathy, amyotrophy, optic atrophy and others [3, 4]. Among them, cerebellar ataxia occurs most frequently in complicated HSP, resulting in “spasticity-ataxia” phenotype [5]. The hereditary modes of HSP include autosomal-dominant (AD), autosomal-recessive (AR), X-linked and maternal trait of inheritance which due to mitochondrial impairment [4]. In all these hereditary modes, AR inheritance is the commonest one [6]. Up to date, a total of 78 loci have been implicated in HSP [5].
Recently, CAPN1 has been identified as a causative gene for spastic paraplegia 76 (SPG76, MIM#616907, NM_005186), a complicated form of HSP [7]. The protein encoded by CAPN1 was calpain-1, which was widely expressed in central nervous system (CNS), has been involved in several important functions of synaptic plasticity, synaptic restructuring, axon maturation and maintenance [8–10]. In 2016, mutations of CAPN1 [c.884G > C (p.R295P), c.1579C > T (p.Q527*), c.406delC (p.P136Rfs*40) and c.1605 + 5G > A] were identified in three AR inherited HSP pedigrees for the first time [7]. Subsequently, other homozygous or compound-heterozygous mutations of CAPN1 were reported in other groups [11–17].
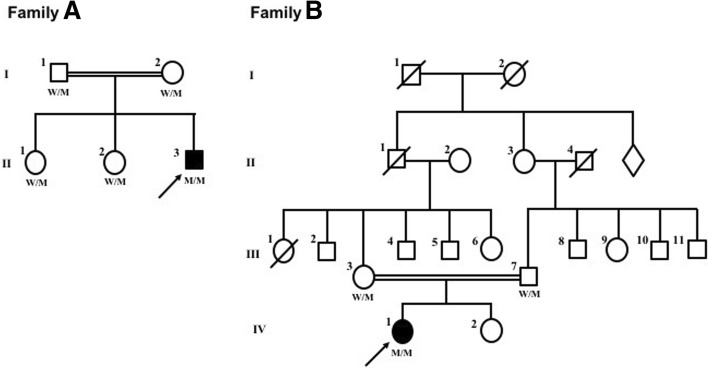
In this study, we reported two Chinese HSP probands, both from consanguineous family (Fig. 1), each carried a novel homozygous mutation of CAPN1. To our knowledge, they were the first SPG76 patients reported in China. Their clinical features and disease progressions were demonstrated retrospectively and would broaden the molecular and clinical spectrum of Chinese HSP patients.
Fig. 1.
Pedigrees of family A and family B with CAPN1 mutations. Arrow: proband; square: male; circle: female; slash: deceased; solid symbol: affected. W/M and M/M represent the genotype of CAPN1. W/M: wild type/mutant; M/M: mutant/mutant
Methods
The probands with walking problems such as spastic or spastic-ataxic gait were collected in the Neurogenetics clinic in Huashan Hospital (Shanghai, PRC). The clinical materials were investigated in both probands.
Genomic DNA was extracted from peripheral blood of both patients and their parents or siblings. Genetic testing of the probands were performed by target sequencing of a panel containing over 4000 known virulence genes. The sequencing was carried out by Illumina HiSeq X-ten platform. The variants screen protocol was as previously reported [18]. The candidate mutations were further confirmed by polymerase chain reaction (PCR) and Sanger sequencing. These mutations were also performed in the parents or siblings to confirm the family co-segregation.
Written informed consents were obtained from both patients and their relatives. This study was approved by the ethics committee of Huashan Hospital.
Results
Results of genetic testing
In family A, the mean depth of target sequencing was 73.5X and the coverage was 100%. The percentage of the target region with mean depth > 20X was 97.0%. According to the screening criteria of low variants frequencies [< 1% in 1000Genome (http://www.1000genomes.org/home), ExAC (http://exac.broadinstitute.org/)] and homozygous mapping, 38 variants were left. But after further screened by clinical manifestations, only one novel homozygous mutation of c.142C > T (p.R48X, NC_000011.10:g.64950314C > T) in CAPN1 (NM_001198868) was found with the depth of 84X.
In family B, the mean depth of target sequencing, the coverage and the percentage of the target region with mean depth > 20X was 107.8X, 99.3 and 97.4% respectively. After screened by the criteria mentioned above, one novel homozygous mutation of c.1015C > T (p.R339X, NC_000011.10:g.64956067 C > T) in CAPN1 was found with the depth of 70X.
Both mutations have been confirmed in the probands and their parents or siblings by Sanger sequencing. The unaffected parents and two unaffected elder sisters of the proband in Family A all carried c.142C > T in the heterozygous state. In family B, the mutation of c.1015C > T was found heterozygous in the unaffected parents of the proband.
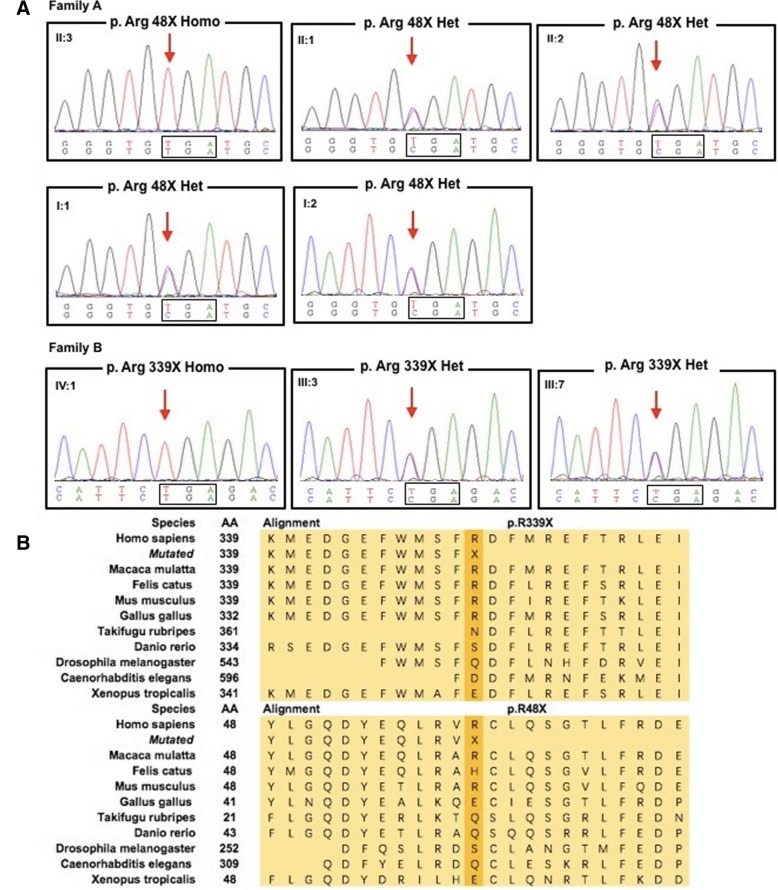
Both variants were partly conserved across species (Fig. 2) and was predicted to be disease causing by mutationtaster (mutationtaster.org) since the truncated mutant took place in positions of R48 and R339 which might cause nonfunctional protein product or affect functional subdomains of the protein.
Fig. 2.
a Sanger sequencing of both probands and their other unaffected family members. b Sequence alignment of calpain-1 across species
Clinical characteristics
The proband from family A gradually developed walking difficulties and stiffness in the lower limbs at age 18. The symptoms deteriorated slowly. He came to our clinic at age 38 and shown a typical spastic gait. Neurologic examination revealed that the muscle tone in both lower limbs was extremely high. Knee and ankle hyperreflexia were also found in both lower limbs. Bilateral Hoffmann sign and Babinski sign were positive. He could still walk along a straight line. Finger-to-nose test and diadochokinesia were performed well. The examination of ocular movement was fine. No distal sensory impairment, cognitive deterioration, bladder dysfunction, or dysarthria was complained. Nerve conduction study and electromyography did not reveal any neurogenic and myogenic damages. The results of head and spinal cord magnetic resonance image (MRI) were also negative.
The proband from family B referred to our clinic for progressive walking difficulties at age 41. Five years ago, she reported weakness in the lower limbs and there was a slight tiptoe when she was walking. She felt slight imbalance and could not walk along a straight line well. The neurological examination revealed hyperreflexia in four limbs and positive bilateral Hoffmann sign. She presented with a moderately spastic-ataxic gait. She had slight bilateral dysmetria when performed finger-to-finger test and mild dysdiadochokinesia. The heel knee test was fine. The ocular pursuit and saccades were normal. She scored 6/40 on the Scale for the Assessment and Rating of Ataxia (SARA). The mini-mental state examination score was 29/30 (education year of 14) suggesting the normal cognitive function. No dysarthria, distal sensory impairment or bladder dysfunction was reported. Neither cerebellum nor spinal cord showed significant atrophy on MRI.
Literatures review
A total of nineteen pedigrees including 35 patients (24 Female, 11 Male patients) with CAPN1 mutations reported till recently were reviewed (Table 1). All the patients showed a pattern of AR inheritance and 85.7% pedigrees were consanguineous. Thirty patients carried homozygous mutations and five patients carried compound heterozygous mutations. The onset age ranged from five to 39 years old. With all the available clinical materials, lower limbs spasticity, presenting with stiffness, hyperreflexia and pathological signs, developed in around 80% patients, followed by cerebellar ataxia developing in 62.9% of the cases, dysarthria in 51.4%, skeletal or tendon deformity in 31.4%. Weakness in lower limbs and ocular movement disorder could also be seen. Some patients developed abnormal bladder function, dysphagia, peripheral neuropathy, intention tremor and even other uncommon symptoms.
Table 1.
Literatures review of reported HSP patients with CAPN1 mutations
| Study (year) | Case No. | Gender | Population | Consanguinity | Mutations | Het/Hom | Exon | Transcript | Age at onset (year) | Clinical features | MR or CT imaging (Brain or spine) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower limbs spasticity | Lower limbs hyperreflexia | Upper limbs hyperreflexia | Babinski sign | Skeletal or tendon deformity | Weakness or amyptrophy | Ocular movement disorder | Abnormal bladder function | Dysarthria | Ataxia | Additional symptoms | |||||||||||
| Gan-Or Z, et al. (2016) [7] | 3 | M | Mornoccan | Y | C.884G > C (p.R295P) | Hom | exon8 | NM_005186 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| F | 20 | + | + | + | + | + | + | – | + | + | – | NA | NA | ||||||||
| F | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | ||||||||
| 5 | M | Mornoccan | Y | C.1579C > T (p.Q527*) | Hom | exon14 | 35 | + | + | + | + | – | + | – | – | + | – | hypoesthesia, peripheral, neuropathy, dysarthria, akinetic face, abolished sympathetic skin reflex in lower limbs | NA | ||
| F | 36 | + | + | + | + | + | + | – | – | + | + | peripheral neuropathy, facial hypokinesia, abolished sympathetic skin reflex in lower limbs | NA | ||||||||
| M | 22 | + | + | + | + | + | + | – | – | + | – | NA | – | ||||||||
| M | 39 | + | + | + | – | – | – | + | – | + | + | NA | NA | ||||||||
| F | 24 | + | + | + | + | + | – | – | – | + | – | abolished sympathetic skin reflex in lower limbs | NA | ||||||||
| 2 | M | Ladho and Utah | N | C.406delC (p.P136Rfs*40) c.1605 + 5 G > A |
Com-het | exon4 exon14 |
33 | + | NA | NA | + | + | NA | – | – | NA | – | NA | mild atrophy of cervical spinal cord | ||
| F | 19 | + | + | + | + | + | + | – | + | NA | + | NA | slightly prominent sulci | ||||||||
| Wang Y, et al. (2016) [11] | 2 | F | Bangladeshi | Y | c.337 + 1 G > A | Hom | exon3 | NM_001198868 | Late teens | + | + | NA | NA | NA | NA | NA | NA | + | + | dysphagia, mild cognitive decline | mild cerebellar atrophy |
| F | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | |||||||||
| 1 | F | Italian | NC | c.183dupC (p.F61 fs) | Hom | exon2 | 25 | + | + | + | + | NA | NA | NA | + | + | + | dysphagia, bilateral positive Hoffmann’s reflex | – | ||
| 2 | F | Tunisian | Y | c.1534C > T (p.R512C) | Hom | exon13 | 23 | + | + | + | NA | – | NA | NA | NA | + | + | NA | cerebellar atrophy | ||
| F | 20 | + | + | + | NA | – | NA | NA | NA | + | + | NA | NA | ||||||||
| 1 | F | French | Y | c.463C > T (p.Q155X) c.1142C > T (p.A381V) |
Com-het | exon5 exon10 |
20 | + | + | + | + | + | + | + | + | + | + | dysphagia, hypokinesia, vibration sense at ankles decreased, bilateral positive Hoffmann’s reflex |
cerebellar atrophy, white matter changes, mild vermian atrophy | ||
| Travaglini,L, et al. (2017) [12] | 1 | M | Italian | N | c.221G > A (p.G74D) c.911C > T (p.T304 M) c.1418G > T (p.R473L) |
Com-het | exon2 exon8 exon13 |
NM_001198868 | 5 | + | + | NA | + | NA | NA | NA | NA | NA | NA | spastic hypertonia | – |
| Tadic V, et al. (2017) [13] | 2 | F | NA | Y | c.759 + 1 G > A | Hom | exon6 | NM_001198868 | 29 | + | + | NA | + | + | NA | + | NA | + | + | muscle hypertonic | cerebellar vermal atrophy |
| F | 33 | NA | + | NA | ± | + | NA | NA | NA | NA | + | slight intention tremor | NA | ||||||||
| Kocoglu C, et al. (2018) [15] | 1 | F | NA | NA | c.994G > A (p.G332R) | Hom | exon9 | NM_001198868 | 21 | + | NA | NA | NA | + | + | NA | NA | + | + | upper limb spasticity keratoconus | – |
| 2 | F | NA | Y | c.1176G > A (P.R392*) | Hom | exon10 | 15 | + | + | NA | + | + | NA | NA | NA | + | + | upper limb spasticity | – | ||
| F | 15 | + | + | NA | + | NA | NA | NA | NA | + | + | NA | NA | ||||||||
| Lambe J, et al. (2018) [14] | 1 | F | Irish | N | c.1534C > T (p.R512C) | Hom | exon13 | NM_001198868 | 14 | + | + | + | + | NA | NA | – | NA | – | + | NA | midbrain pons, cerebellar atrophy, spinal cord normal |
| Shetty A, et al. (2018) [16] | 1 | F | Japanese | Y | c.2118 + 1G > T | Hom | exon21 | NA | 37 | NA | NA | NA | NA | NA | NA | NA | NA | NA | + | upper motor neuron findings in the legs | NA |
| 2 | M | Turkish | Y | c.397C > T | Hom | exon4 | NA | 23 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | progressive spastic paraparesis | NA | |
| F | NA | 20 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | severe proximal weakness | NA | |||||||
| 1 | M | Punjabi | Y | c.843 + 1G > C | Hom | exon7 | NA | 37 | + | NA | NA | NA | NA | NA | NA | NA | NA | + | spastic quadriparesis | NA | |
| Melo US. et al. (2018) [17] | 3 | F | Brazilian | Y | c.1176G > A (P.R392*) | Hom | exon10 | NM_001198868 | NA | + | NA | NA | NA | NA | NA | NA | NA | + | two of three | NA | NA |
| F | NA | + | NA | NA | NA | NA | NA | NA | NA | + | NA | NA | |||||||||
| M | NA | + | NA | NA | NA | NA | NA | NA | NA | + | NA | NA | |||||||||
| 1 | F | Y | c.1176G > A (P.R392*) | Hom | exon10 | 22 | + | + | NA | NA | NA | NA | NA | NA | NA | – | NA | NA | |||
| 2 | F | Y | c,675C > A p.Y225* | Hom | eoxn6 | 20 | + | NA | NA | NA | NA | NA | NA | NA | NA | + | NA | NA | |||
| M | 35 | + | NA | NA | NA | NA | NA | NA | NA | NA | + | NA | NA | ||||||||
| 2 | F | N | c.1176G > A (P.R392*) c.618_619 delAG (p.G208 Qfs*7) |
Com-het | exon10 exon6 |
30 | + | NA | NA | NA | NA | NA | NA | NA | NA | + | NA | NA | |||
| M | Y | c.1176G > A (p.R392*) | Hom | exon10 | 38 | + | NA | NA | NA | NA | NA | NA | NA | NA | + | NA | NA | ||||
| Current study | 2 | M | Chinese | Y | c.142C > T (p.R48*) | Hom | exon2 | NM_001198868 | 18 | + | + | + | + | – | – | – | NA | – | – | muscle hypertonic in lower limbs, bilateral positive Hoffmann’s reflex | – |
| F | Y | c.1015C > T (p.R339X) | Hom | exon10 | 41 | + | + | + | – | – | + | – | – | – | + | bilateral positive Hoffmann’s reflex | – | ||||
AR autosomal-recessive, com-het compound heterozygous, CT computed tomography, hom homozygous, HSP Hereditary Spastic Paraplegia, F female, M male, MRI magnetic resonance image, N no, NA not available, NC not certain, Y yes, y years old, +: positive, −: negative or normal, ±:suspicious
Discussion
With the wildly application of next generation sequencing, more and more classical “HSP genes” causing cerebellar ataxia were found and vice versa. So, these genes could be categorized as “spasticity-ataxia” spectrum. According to a review in 2017, genes related to “spasticity-ataxia” spectrum was expanded to 69 members [5]. CAPN1 was one of them, manifesting as pure HSP or complicated HSP. The mutations in CAPN1 causing autosomal recessive HSP have been found since 2016 by whole exome sequencing in three pedigrees. Among these patients carrying CAPN1 mutations, lower limbs spasticity was the predominant symptom combined with cerebellar ataxia or not. Therefore, “spasticity-ataxia” phenotype might conduce to the diagnosis of SPG76.
The protein product of CAPN1, calpain-1, also known as μ-calpain, contains four domains: the N-terminal anchor helix region, the CysPc protease domain (including two protease core domains of PC1 and PC2), the C2 domain-like domain and the penta-EF-hand domain (PEF). As a calcium-activated cysteine protease, calpain-1 binds to calcium through PEF domain [19]. It has been proved that the activation of calpain-1 is required for its neuro-protective role in CNS [20]. Several mechanisms for the protective role were suggested by interacting with CDK5 and NR2B to control NMDA-receptor degradation [21] or affecting AMPA receptors through degradation of its substrate, glutamate receptor-interacting protein [22]. In calpain-1 deficient mice, dysfunction of calpain-1 reduced dendritic branching complexity and led to spine density deficits [23]. In zebrafish embryos, knockdown of calpain-1 induced disruption of microtubule network in brain and spinal cord [24], which indicated that dysfunction of calpain-1 could result in neurodegeneration or disorganization of neurons [25]. Immunohistochemistry study revealed that calpain-1 was the major calpain isoform in cerebellar neurons [26], and the activity of it in cerebellum was higher than that in cortex or hippocampus [27], suggesting that calpain-1 played a key role in maintaining the normal cerebellar function.
All the reported mutations scattered in exons 2–6, 8–10, 13, 14 and 21of CAPN1, potentially damaged the normal structure of calpain-1 or led to early termination of protein coding, causing the dysfunction of calpain-1. In this current study, the probands were in accordance with two HSP subtypes: the male patient presented with pure HSP subtype with normal cerebellar function, while the female patient manifested as classical complicated HSP subtype showing symptoms of both HSP and cerebellar ataxia. Two novel homozygous mutations c.1176G > A and c.675C > A of CAPN1 were detected respectively. These two mutations were situated in exon 2 and exon 10 and brought on a premature stop codon at the positions of R48 and R339, causing the destruction of calpain-1 normal structure. The structural incompleteness of calpain-1 would interfere with its neuro-protective role in CNS and induce neurodegeneration or disorganization of neurons, which might lead to SPG76.
Conclusion
Together with previously reported cases, our study broadened the clinical and molecular spectrum of CAPN1-related SPG76 and exemplified the concept of “spasticity-ataxia” phenotype, further increasing our understanding of complicated HSP form and its connection with cerebellar ataxia. All these observations indicated that CAPN1 screening is necessary in HSP patients, especially when patients suffer from spasticity-ataxia phenotype.
Acknowledgements
We sincerely appreciate the patients and their relatives for their help and willingness in this study.
Funding
This work was supported by grants from the National Natural Science Foundation of China(No.81401048).
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Abbreviations
- AD
Autosomal-dominant
- AR
Autosomal-recessive
- CNS
Central nervous system
- HSP
Hereditary spastic paraplegias
- MRI
Magnetic resonance image
- PCR
Polymerase chain reaction
- PEF
Penta-EF-hand domain
- SARA
Scale for the Assessment and Rating of Ataxia
- SPG76
Spastic paraplegia 76
Authors’ contributions
FP performed the statistical analysis and drafted the manuscript. YS carried out the molecular genetic studies and drafted the manuscript. CQ participated in the design of the study. JW participated in the design of the study. JW conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the ethics committee of Huashan Hospital. Written informed consents for participation were obtained from both patients and their relatives.
Consent for publication
Written informed consents for publication were obtained from both patients and their relatives.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harding AE. Hereditary "pure" spasticparaplegia: a clinical and genetic study of 22 families. J Neurol Neurosurg Psychiatry. 1981;44(10):871–883. doi: 10.1136/jnnp.44.10.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A. Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Expl Neurol. 2014;261:518–539. doi: 10.1016/j.expneurol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Finsterer J, Löscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci. 2012;318(1–2):1–18. doi: 10.1016/j.jns.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 5.Synofzik M, Schule R. Overcoming the divide between ataxias and spastic paraplegias: shared phenotypes, genes, and pathways. Mov Disord. 2017;32(3):332–345. doi: 10.1002/mds.26944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reid E. Pure hereditary spastic paraplegia. J Med Genet. 1997;34(6):499–503. doi: 10.1136/jmg.34.6.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gan-Or Z, Bouslam N, Birouk N, Lissouba A, Chambers DB, Veriepe J, et al. Mutations in CAPN1 cause autosomal-recessive hereditary spastic paraplegia. Am J Hum Genet. 2016;98(5):1038–1046. doi: 10.1016/j.ajhg.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zadran S, Jourdi H, Rostamiani K, Qin Q, Bi X, Baudry M. Brain-derived neurotrophic factor and epidermal growth factor activate neuronal m-Calpain via mitogen-activated protein kinase-dependent phosphorylation. J Neurosci. 2010;30(3):1086–1095. doi: 10.1523/JNEUROSCI.5120-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Liu MC, Wang KK. Calpain in the CNS: from synaptic function to neurotoxicity. Sci Signal. 2008;1(14):re1. doi: 10.1126/stke.114re1. [DOI] [PubMed] [Google Scholar]
- 10.Goll DE, Thompson VF, Li H, Wei W, Cong J. The Calpain system. Physiol Rev. 2003;83(3):731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Hersheson J, Lopez D, Hammer M, Liu Y, Lee KH, et al. Defects in the CAPN1 gene result in alterations in cerebellar development and cerebellar Ataxia in mice and humans. Cell Rep. 2016;16(1):79–91. doi: 10.1016/j.celrep.2016.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Travaglini L, Bellacchio E, Aiello C, Pro S, Bertini E, Nicita F. Expanding the clinical phenotype of CAPN1-associated mutations: a new case with congenital-onset pure spastic paraplegia. J Neurol Sci. 2017;378:210–212. doi: 10.1016/j.jns.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Tadic V, Klein C, Hinrichs F, Munchau A, Lohmann K, Bruggemann N. CAPN1 mutations are associated with a syndrome of combined spasticity and ataxia. J Neurol. 2017;264(5):1008–1010. doi: 10.1007/s00415-017-8464-5. [DOI] [PubMed] [Google Scholar]
- 14.Lambe J, Monaghan B, Munteanu T, Redmond J. CAPN1 mutations broadening the hereditary spastic paraplegia/spinocerebellar ataxia phenotype. Pract Neurol. 2018;18(5):369–372. doi: 10.1136/practneurol-2017-001842. [DOI] [PubMed] [Google Scholar]
- 15.Kocoglu C, Gundogdu A, Kocaman G, Kahraman-Koytak P, Uluc K, Kiziltan G, et al. Homozygous CAPN1 mutations causing a spastic-ataxia phenotype in 2 families. Neurol Genet. 2018;4(1):e218. doi: 10.1212/NXG.0000000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shetty A, Ashtiani S, Gan-Or Z, Van de Warrenburg B, Wassenberg T, Rouleau G, et al. CAPN1: novel mutations expanding the phenotype of hereditary spastic paraparesis. (P6.039). Eur J Med Genet. 2018;90(15 Supplement). https://www.ncbi.nlm.nih.gov/pubmed/30572172. [DOI] [PubMed]
- 17.Melo US, Freua F, Lynch DS, Ripa BD, Tenorio RB, Saute JAM, et al. Clinical aspects of hereditary spastic paraplegia 76 and novel CAPN1 mutations. Clin Genet. 2018;94(5):482–483. doi: 10.1111/cge.13428. [DOI] [PubMed] [Google Scholar]
- 18.Chen K, Yang K, Luo SS, Chen C, Wang Y, Wang YX, et al. A homozygous missense variant in HSD17B4 identified in a consanguineous Chinese Han family with type II Perrault syndrome. BMC Med Genet. 2017;18(1):91. doi: 10.1186/s12881-017-0453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono Y, Sorimachi H. Calpains: an elaborate proteolytic system. Biochim Biophys Acta. 2012;1824(1):224–236. doi: 10.1016/j.bbapap.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Baudry M, Bi X. Calpain-1 and Calpain-2: the yin and Yang of synaptic plasticity and neurodegeneration. Trends Neurosci. 2016;39(4):235–245. doi: 10.1016/j.tins.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawasli AH, Benavides DR, Nguyen C, Kansy JW, Hayashi K, Chambon P, et al. Cyclin-dependent kinase 5 governs learning and synaptic plasticity via control of NMDAR degradation. Nat Neurosci. 2007;10(7):880–886. doi: 10.1038/nn1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X, Wyszynski M, Sheng M, Baudry M. Proteolysis of glutamate receptor-interacting protein by calpain in rat brain: implications for synaptic plasticity. J Neurochem. 2001;77(6):1553–1560. doi: 10.1046/j.1471-4159.2001.00359.x. [DOI] [PubMed] [Google Scholar]
- 23.Amini M, Ma CL, Farazifard R, Zhu G, Zhang Y, Vanderluit J, et al. Conditional disruption of calpain in the CNS alters dendrite morphology, impairs LTP, and promotes neuronal survival following injury. J Neurosci. 2013;33(13):5773–5784. doi: 10.1523/JNEUROSCI.4247-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HL, Yuh CH, Wu KK. Nestin is essential for zebrafish brain and eye development through control of progenitor cell apoptosis. PLoS One. 2010;5(2):e9318. doi: 10.1371/journal.pone.0009318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Briz V, Chishti A, Bi X, Baudry M. Distinct roles for mu-calpain and m-calpain in synaptic NMDAR-mediated neuroprotection and extrasynaptic NMDAR-mediated neurodegeneration. J Neurosci. 2013;33(48):18880–18892. doi: 10.1523/JNEUROSCI.3293-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamakubo T, Kannagi R, Murachi T, Matus A. Distribution of calpains I and II in rat brain. J Neurosci. 1986;6(11):3103–11. doi: 10.1523/JNEUROSCI.06-11-03103.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baudry M, Simonson L, Dubrin R, Lynch G. A comparative study of soluble calcium- dependent proteolytic activity in brain. J Neurobiol. 1986;17(1):15–28. doi: 10.1002/neu.480170103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.