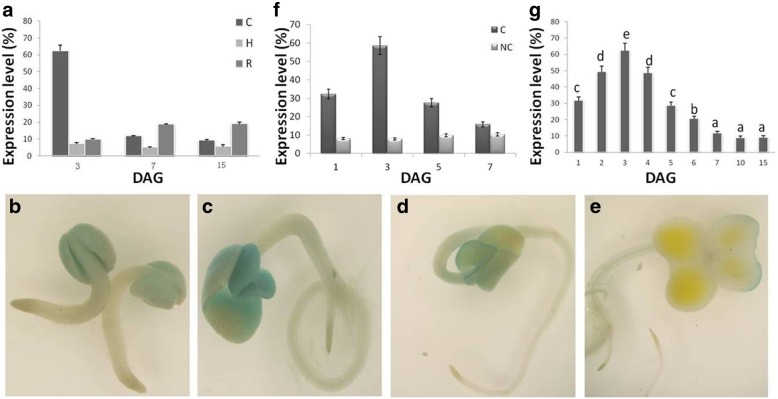
Fig. 1.
Expression profiling of BnPHT1;4 in early developing seedlings of Brassica napus. (a) Quantitative RT-PCR analysis of BnPHT1;4 expression in oilseed rape seedlings at 3–15 DAG. Total RNA was isolated from roots (R), hypocotyls (H), cotyledons (C) of seedlings cultured in normal conditions (1 mM Pi) at 3, 7 and 15 DAG for RT-PCR analysis, using BnACT2 gene as standard control. The error bars indicate the standard errors (n = 3). (b-e). Histochemical assay of GUS activity in early developing seedlings. GUS staining observation of transgenic B. napus plants harboring the BnPHT1;4 promoter: GUS fusion gene in 1 DAG (b), 3 DAG (c), 5 DAG (d) and 7 DAG (e) seedlings cultured on MS medium with 1 mM Pi, respectively. (f) Quantitative RT-PCR analysis of the GUS gene in cotyledons (C) and in roots plus leaves and hypocotyls (NC) of BnPHT1;4 promoter: GUS transgenic plants. Total RNAs were isolated from C and NC of 1 DAG, 3 DAG, 5 DAG and 7 DAG seedlings cultured on MS medium with 1 mM Pi, respectively, for RT-PCR analysis, using BnACT2 gene as standard control. (g) Quantitative RT-PCR analysis of BnPHT1;4 expression in cotyledons of oilseed rape seedlings at 1–15 DAG. Total RNAs was isolated from cotyledons of seedlings cultured in normal conditions (1 mM Pi) at 1–15 DAG for RT-PCR analysis, using BnACT2 gene as standard control. The error bars indicate the standard errors (n = 9). Significance of difference was analyzed by Duncan’s test (P < 0.05). Different lowercase letters indicate statistically significant differences. DAG, day after germination