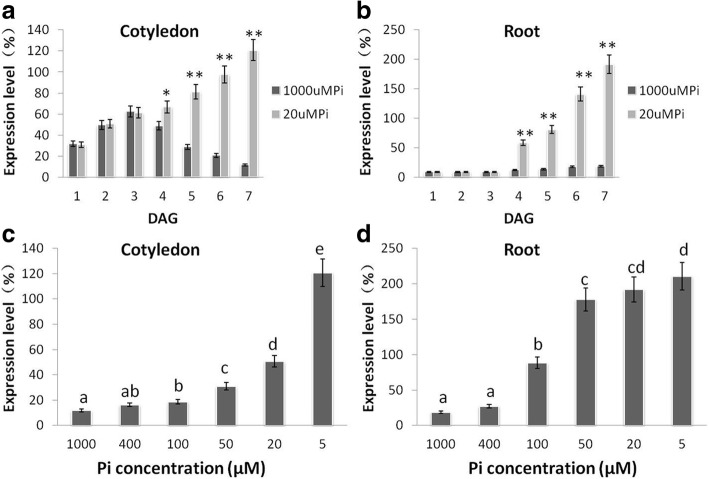
Fig. 2.
Expression profiling of BnPHT1;4 in cotyledons and roots of Brassica napus under Pi starvation. (a and b) Quantitative RT-PCR analysis of BnPHT1;4 expression in cotyledons (a) and roots (b) of oilseed rape seedlings at 1–7 DAG. Total RNA was isolated from cotyledons (a) and roots (b) of seedlings under 1000 or 20 μM Pi at 1–7 DAG for RT-PCR analysis, using BnACT2 gene as standard control. (c and d) Quantitative RT-PCR analysis of BnPHT1;4 expression in cotyledons (c) and roots (d) of oilseed rape seedlings under different Pi concentrations. The 7 DAG seedlings were transferred to 5, 20, 50, 100, 400 or 1000 μM Pi for 3 days, and then total RNA was isolated from cotyledons (c) and roots (d) for RT–PCR analysis, using BnACT2 gene as standard control. KCl was used to replace KH2PO4 in the medium for the equivalent amount of potassium. The error bars indicate the standard errors. Significance of difference was analyzed by Duncan’s test (*p<0.05, **p<0.01, n = 3). Different lowercase letters indicate statistically significant differences. DAG, day after germination