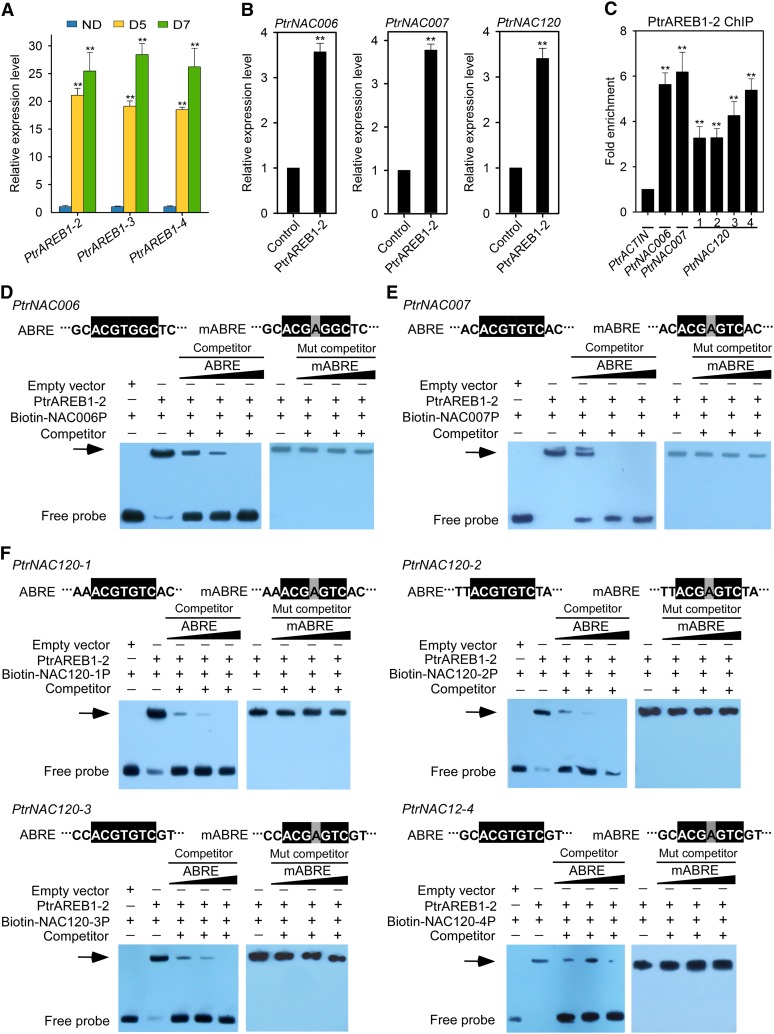
Figure 5.
PtrAREB1-2 Activates the Transcription of PtrNAC Genes and Binds Directly to the ABRE Motifs in Their Promoters.
(A) Expression patterns of PtrAREB1-2, PtrAREB1-3, and PtrAREB1-4 genes in response to drought stress detected by RT-qPCR. Expression was highly induced by drought treatment. Error bars indicate 1 se of three biological replicates from independent pools of P. trichocarpa SDX tissues. Asterisks indicate significant differences between control (ND) and drought-treated (D5 and D7) samples for each gene (**, P < 0.01, Student’s t test).
(B) RT-qPCR to detect the transcript abundance of PtrNAC006, PtrNAC007, and PtrNAC120 in SDX protoplasts overexpressing GFP (Control) or PtrAREB1-2 in the presence of external 50 µM ABA. Control values were set as 1. Error bars indicate 1 se of three biological replicates (three independent batches of SDX protoplast transfections). Asterisks indicate significant differences for each gene between control protoplasts and those overexpressing PtrAREB1-2 samples for each gene (**, P < 0.01, Student’s t test).
(C) PtrAREB1-2 ChIP assays showing that PtrAREB1-2 binds directly to the promoters of PtrNAC genes. P. trichocarpa SDX protoplasts overexpressing PtrAREB1-2-GFP or GFP (control) were used for the ChIP assay with anti-GFP antibody, and the precipitated DNA was quantified by qPCR. Enrichment of DNA was calculated as the ratio between 35Spro:PtrAREB1-2-GFP and 35Spro:GFP (control), normalized to that of the PtrACTIN gene. Numbers indicate ABRE motif sites in PtrNAC120. Error bars represent 1 se of three biological replicates (three independent batches of SDX protoplast transfections). Asterisks indicate significant differences between the control fragment (PtrACTIN) and each fragment containing an ABRE motif (**, P < 0.01, Student’s t test).
(D) to (F) Nucleotide sequences of the wild-type ABRE and a mutated ABRE motif (mABRE) (top panels). Core sequences are shaded in black, and the mutated nucleotide is shaded in gray. EMSA analysis of PtrAREB1-2 binding to ABRE motifs in PtrNAC006 (D), PtrNAC007 (E), and PtrNAC120 (F) promoters is shown in the bottom panels. The arrows show the shifted band representing the protein-DNA complex. PtrNAC006 (D), PtrNAC007 (E), and PtrNAC120 (F) promoter fragments were labeled with biotin. Fragments without biotin labeling were used as competitors. Wild-type or mutated ABRE competitors were used in a molar excess of 50×, 100×, or 150×.