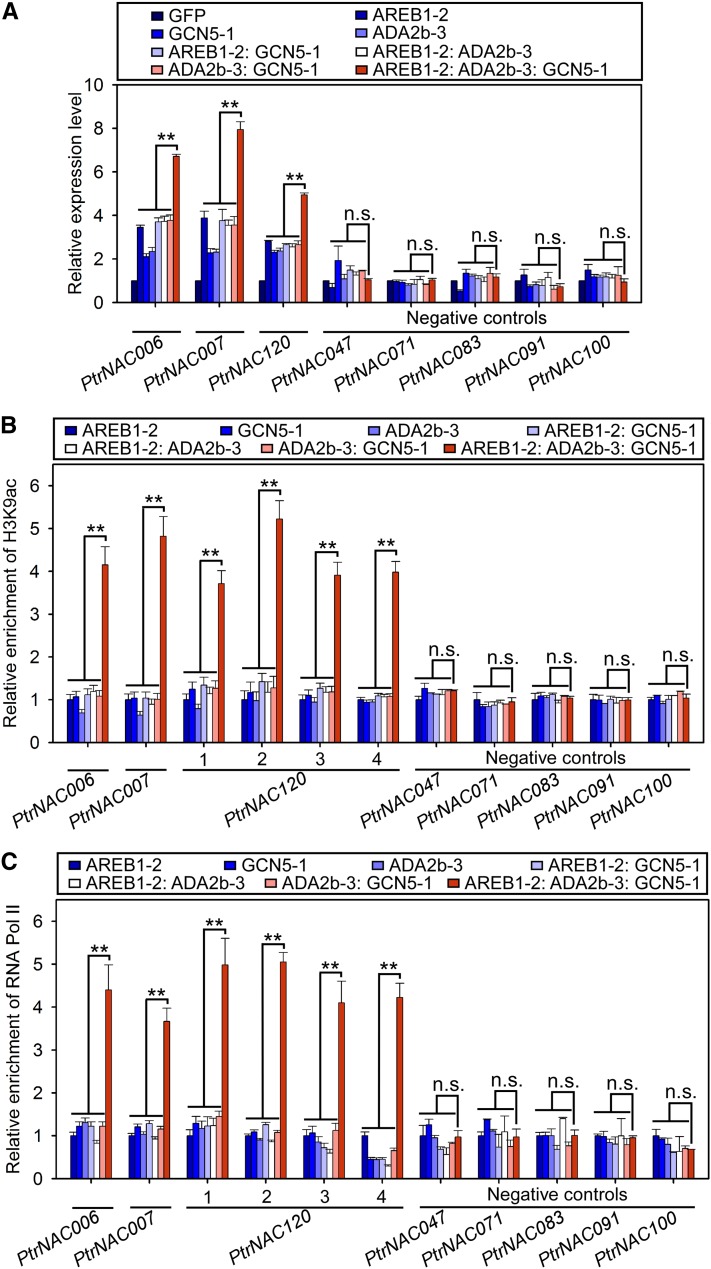
Figure 7.
PtrADA2b-3 and PtrGCN5-1 Together Enhance PtrAREB1-2-Mediated Transcriptional Activation of PtrNAC Genes by Increasing H3K9ac Level and RNA Pol II Recruitment at Their Promoters.
(A) RT-qPCR detection of PtrNAC006, PtrNAC007, and PtrNAC120 transcripts in P. trichocarpa SDX protoplasts overexpressing GFP (control), PtrAREB1-2, PtrGCN5-1, PtrADA2b-3, PtrAREB1-2:PtrGCN5-1, PtrAREB1-2:PtrADA2b-3, PtrADA2b-3:PtrGCN5-1, or PtrAREB1-2:PtrADA2b-3:PtrGCN5-1 in the presence of external 50 µM ABA. Five genes without ABRE motifs were used as negative controls, none of which had activated expression. The control values were set as 1. Error bars represent 1 se of three biological replicates (three independent batches of SDX protoplast transfections). Asterisks indicate significant differences between the ternary complex and each monomeric or dimeric protein for each gene (**, P < 0.01, Student’s t test), and n.s. denotes no significant difference.
(B) and (C) ChIP-qPCR showing that co-overexpression of PtrAREB1-2, PtrADA2b-3, and PtrGCN5-1 increased H3K9ac (B) and RNA Pol II (C) enrichment at the promoters of PtrNAC006, PtrNAC007, and PtrNAC120. SDX protoplasts overexpressing PtrAREB1-2, PtrGCN5-1, PtrADA2b-3, PtrAREB1-2:PtrGCN5-1, PtrAREB1-2:PtrADA2b-3, PtrADA2b-3:PtrGCN5-1, PtrAREB1-2:PtrADA2b-3:PtrGCN5-1, or GFP (control) were used for ChIP assays with anti-H3K9ac (B) and anti-RNA Pol II (C) antibodies, and the precipitated DNA was quantified by qPCR. None of the five negative control genes had enhanced H3K9ac (B) or RNA Pol II (C) levels at their promoters. Enrichment values represent the relative fold change compared with the protoplasts overexpressing PtrAREB1-2. Error bars indicate 1 se of three biological replicates (three independent batches of SDX protoplast transfections). Asterisks indicate significant differences between the ternary complex and each monomeric or dimeric protein for each fragment containing the ABRE motif (**, P < 0.01, Student’s t test), and n.s. denotes no significant difference.