Summary
The mechanisms that restrict peptidoglycan biosynthesis to the pole during elongation and re-direct peptidoglycan biosynthesis to mid-cell during cell division in polar-growing Alphaproteobacteria are largely unknown. Here, we explore the role of early division proteins of Agrobacterium tumefaciens including 3 FtsZ homologs, FtsA, and FtsW in the transition from polar growth to mid-cell growth and ultimately cell division. Although two of the three FtsZ homologs localize to mid-cell, exhibit GTPase activity and form co-polymers, only one, FtsZAT, is required for cell division. We find that FtsZAT is required not only for constriction and cell separation, but also for initiation of peptidoglycan synthesis at mid-cell and cessation of polar peptidoglycan biosynthesis. Depletion of FtsZAT in A. tumefaciens causes a striking phenotype: cells are extensively branched and accumulate growth active poles through tip splitting events. When cell division is blocked at a later stage by depletion of FtsA or FtsW, polar growth is terminated and ectopic growth poles emerge from mid-cell. Overall, this work suggests that A. tumefaciens FtsZ makes distinct contributions to the regulation of polar growth and cell division.
Keywords: Agrobacterium, polar elongation, FtsZ, bacterial cell division, peptidoglycan
Graphical Abstract
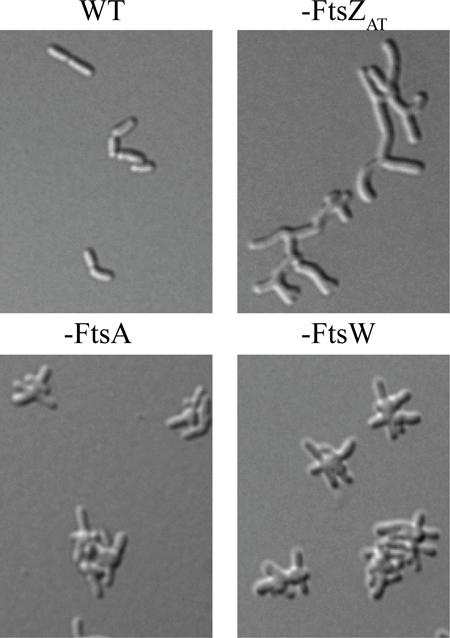
In Agrobacterium tumefaciens unipolar elongation is followed by growth at mid-cell, enabling cell division. Remarkably, the absence of FtsZAT causes growth poles to accumulate due to tip splitting events because peptidoglycan synthesis is not redirected from the growth pole to mid-cell. In contrast, the absence of downstream division proteins, FtsA or FtsW, causes ectopic growth poles to emerge from mid-cell, indicating that these proteins are not necessary for the redirection of growth to mid-cell.
Introduction
The spatial and temporal regulation of cell division is a vital process across bacterial species with implications in the development of antimicrobial therapies (den Blaauwen et al., 2014). The cell division process must coordinate membrane invagination(s), peptidoglycan (PG) biosynthesis and remodeling, and the physical separation of the two daughter cells, all while maintaining cellular integrity. Furthermore, cell division must be precisely regulated to be orchestrated with other key cell cycle processes including cell elongation, DNA replication, and chromosome segregation to ensure that each daughter cell is of sufficient size and contains a complete genome (Haeusser & Levin, 2008, Wu & Errington, 2004).
To initiate bacterial cell division, the tubulin-like GTPase, FtsZ, polymerizes and forms a discontinuous ring-like structure at the future site of cell division (Bi & Lutkenhaus, 1991, de Boer et al., 1992, Li et al., 2007, Fu et al., 2010, Holden et al., 2014, Bisson-Filho et al., 2017, Yang et al., 2017). The presence of FtsZ at mid-cell leads to the recruitment of many proteins that function in cell division, collectively called the divisome (Du & Lutkenhaus, 2017, Meier & Goley, 2014, Goley et al., 2011, Erickson et al., 2010). The divisome includes enzymes required for septal PG biosynthesis, such as penicillin-binding protein 3 (PBP3) and FtsW (Du & Lutkenhaus, 2017). Once the divisome is fully assembled, FtsZ filaments treadmill along the circumference of the mid-cell, driving the Z-ring constriction (Yang et al., 2017, Bisson-Filho et al., 2017). The movement of FtsZ filaments is correlated with the movement of enzymes that function in septal PG biogenesis. These finding are consistent with the notion that FtsZ not only recruits enzymes that function in PG biogenesis to mid-cell but also regulates their activities to promote proper cell wall biogenesis (Varma & Young, 2004, Aaron et al., 2007, Sundararajan et al., 2015).
In most rod-shaped model organisms used to study cell division, a block in cell division leads to the production of long, smooth filamentous cells. This phenotype suggests that assembly or activation of some divisome components is necessary not only to enable the cells to divide but also to stop cellular elongation. Indeed, in Escherichia coli, FtsZ (along with the Z-ring stabilizing proteins FtsA, ZipA, and ZapA) has been proposed to have an early function in the switch from lateral PG biogenesis to mid-cell PG biosynthesis (Aarsman et al., 2005). Following maturation of the divisome by recruitment of additional PG remodeling enzymes and cell division proteins, PG biosynthesis is coordinated with membrane invagination, enabling cells to constrict and separate (Gray et al., 2015).
Conversely to e.g. E. coli, polar growing rods in the alphaproteobacterial clade Rhizobiales exhibit branched morphologies when cell division is blocked (Pini et al., 2015, Howell & Brown, 2016, Pini et al., 2013, Bellefontaine et al., 2002, Cheng et al., 2007, Latch & Margolin, 1997, Fujiwara & Fukui, 1974, Zupan et al., 2013). Examination of the cell morphologies resulting from the block in cell division suggests that different types of branched morphologies arise (Figueroa-Cuilan & Brown, 2018). Drug treatments that block DNA replication cause an early block in cell division, resulting in a “Y” morphology in which the branches are formed from existing growth poles (Latch & Margolin, 1997, Fujiwara & Fukui, 1974). In contrast, antibiotics that target PBP3 cause mid-cell bulges and branches with some cells adopting a “T” or “+” morphologies (Latch & Margolin, 1997, Zupan et al., 2013). These observations suggest that polar-like PG synthesis is redirected to mid-cell when cell division is blocked at a later stage. The manifestation of two distinct phenotypes during early and late blocks in cell division suggests that divisome assembly and activation may contribute to termination of polar growth, onset of mid-cell PG biosynthesis, cell constriction, and ultimately cell separation.
In Agrobacterium tumefaciens, homologs of FtsZ and FtsA fused to fluorescent proteins localize at the growth pole during elongation and at mid-cell during division (Brown et al., 2012, Cameron et al., 2014, Zupan et al., 2013). FtsZ was found to arrive at mid-cell considerably earlier than FtsA (Cameron et al., 2014), indicating that FtsZ may be able to initiate Z-ring formation prior to FtsA recruitment to the divisome. This observation is consistent with the described order of divisome assembly in Caulobacter crescentus (Goley et al., 2010) and suggests a distinctive time-dependent role of these proteins in cell division.
Here, we take advantage of the ability to deplete essential proteins in A. tumefaciens (Figueroa-Cuilan et al., 2016) to explore the function of cell division proteins FtsZ, FtsA, and FtsW in a polar growing alphaproteobacterium. Although the genome of A. tumefaciens encodes three FtsZ homologs, we find that only one, henceforth referred to as FtsZAT, is essential for cell survival. FtsZAT is required to recruit division proteins to mid-cell and likely regulates the activity of PG biosynthesis enzymes at mid-cell. In the absence of FtsZAT, cells not only fail to divide but are also unable to terminate polar growth. Depletion of either FtsA or FtsW also causes a block in cell division, but unlike FtsZAT depletion, growth at the poles is halted and instead, polar-like PG synthesis is redirected to mid-cell. These observations suggest that only FtsZAT is required to initiate cell division-specific PG biosynthesis at mid-cell, whereas FtsA and FtsW are exclusively required for cell division. Together these findings suggest that A. tumefaciens uses sequential regulation of cell division to ensure that initiation of growth at mid-cell is followed by constriction and ultimately cell separation, a theme that is broadly conserved in bacteria.
Results and Discussion
FtsZAT is required for cell division and termination of polar growth.
Agrobacterium tumefaciens contains three homologs of Escherichia coli’s FtsZ, Atu_2086, Atu_4673, and Atu_4215 (Figure 1A) (Zupan et al., 2013). E. coli FtsZ comprises three regions: the conserved N-terminal tubulin-like GTPase domain, a C-terminal linker (CTL), and a conserved C-terminal peptide (CTP), which anchors FtsZ to the membrane via interactions with FtsA (Ortiz et al., 2016). Atu_2086 contains each of these domains, out of which the GTPase domain and CTP share 52% and 67% identity to their respective domain in E. coli FtsZ, whereas the CTL is extended in length (Zupan et al., 2013). The gene encoding Atu_2086 is found in a putative operon with genes encoding DdlB, FtsQ, FtsA (Wood et al., 2001, Goodner et al., 2001) and is predicted to be essential for cell survival based on a saturating transposon mutagenesis screen (Curtis & Brun, 2014). Atu_2086 localizes to mid-cell in wildtype (WT) pre-divisional cells (Figure 1B) (Brown et al., 2012, Zupan et al., 2013); consistent with a role in cell division. Atu_4673 (called FtsZ1; consistent with the genome annotation) contains a complete GTPase domain with 49% identity to tubulin domain of E. coli FtsZ but lacks both the CTL and CTP (Zupan et al., 2013). Although Atu_4673 is not predicted to be required for cell survival based on saturating transposon mutagenesis (Curtis & Brun, 2014), it localizes to mid-cell in pre-divisional cells, suggesting a possible role in cell division (Figure 1B). Atu_4215 (termed FtsZ3 in this work) contains a partial GTPase domain with 48% identity to the N-terminal portion of the E. coli FtsZ tubulin domain and lacks both the CTL and CTP (Zupan et al., 2013). FtsZ3 is not essential for survival of A. tumefaciens based on saturating transposon mutagenesis (Curtis & Brun, 2014) and exhibits a diffuse localization pattern (Figure 1B). Together, these data suggest that Atu_2086 is the canonical FtsZ protein required for cell division, and this protein will be referred to as FtsZAT throughout this work (although it is annotated as FtsZ2 in the A. tumefaciens C58 genome (Goodner et al., 2001, Wood et al., 2001)).
Figure 1. Characterization of FtsZ homologs in A. tumefaciens.
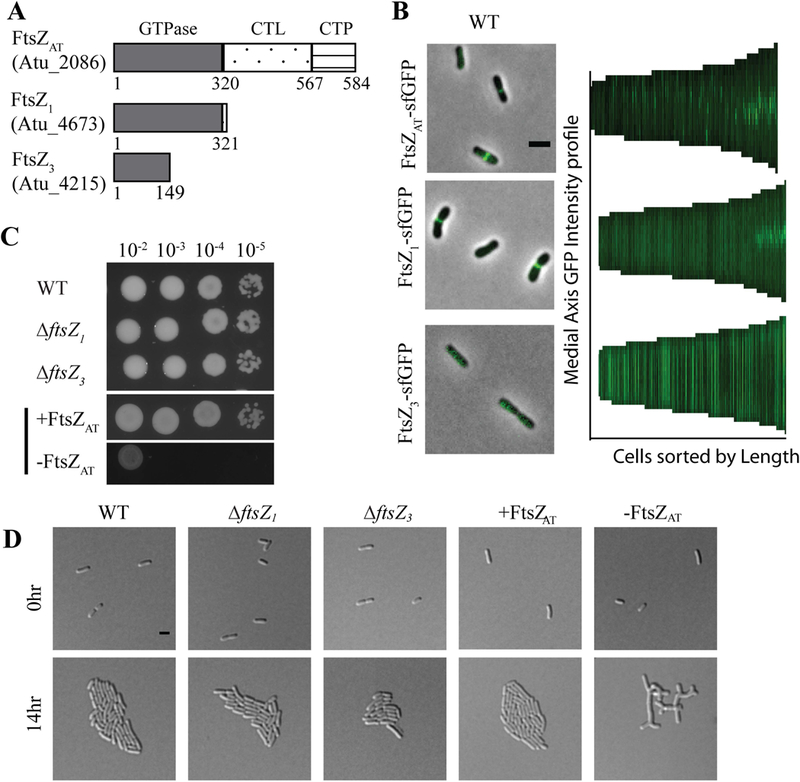
A) Domain schematic of FtsZ homologs in A. tumefaciens. Note that domains are not drawn to scale. B) Representative image of localization patterns for each FtsZ homolog expressed in wildtype cells. Demographs depict localization of FtsZ homologs at a population level. Median profiles of the GFP channel of 225–240 cells per strain are stacked and ordered by cell length. C) Cell viability is shown by spotting serial dilutions. (+FtsZAT is spotted on ATGN with IPTG while all other strains are spotted on ATGN without IPTG). D) Cell morphology and microcolony formation are shown for WT, ∆ftsZ1 and ∆ftsZ3, and the ftsZAT depletion strain under induced (+FtsZAT) and uninduced (-FtsZAT) conditions. All scale bars are set to 2 µm.
To characterize the function of each FtsZ homolog, we constructed deletions of ftsZ1 and ftsZ3 and a depletion strain of ftsZAT. Since we were unable to construct a deletion of ftsZAT, we used a depletion strategy in which ftsZAT is present as a single copy under the control of an isopropyl β-D-1-thiogalactopyranoside (IPTG) inducible promoter at a neutral site in the chromosome (Figueroa-Cuilan et al., 2016, Howell & Brown, 2016). Using western blot analysis, we have confirmed the depletion of FtsZAT in the absence of IPTG (Supplemental Figure 1A).
Deletion of ftsZ1 or ftsZ3 does not impact cell viability (Figure 1C), cell morphology (Figure 1D; Tables 1; Supplemental Figure 1B), microcolony formation (Figure 1D), constriction rate or position (Table 1) when compared to WT cells. Similarly, when FtsZAT is expressed via IPTG induction in the depletion strain (labeled in figures as +FtsZAT) the cells remain viable (Figure 1C), are similar in size to WT cells (Table 1), properly position constrictions (Table 1), and form microcolonies (Figure 1D). In contrast, depletion of FtsZAT (labeled in figures as –FtsZAT) causes a marked decrease in cell viability (Figure 1C) and triggers the formation of large cells with complex branched morphologies (Table 1; Figure 1D). To quantify changes in morphology during depletion of FtsZAT, the cell area of at least 100 cells was calculated based on phase contrast images of cells acquired immediately after removal of the inducer (-FtsZAT 0 h), 8 h after removal of the inducer (-FtsZAT 8 h), and 14 h after removal of the inducer (-FtsZAT 14 h) (Table 1, Supplemental Figure 1C). Initially, the FtsZAT depleted cells are similar to WT in cell size, but after 8 h of FtsZAT depletion the cell area has nearly doubled (Table 1, Supplemental Figure 1C). Within 14 h of FtsZAT depletion, the average cell area has dramatically increased (Table 1, Supplemental Figure 1C). Together, these results demonstrate that only the FtsZAT homolog is required for proper cell growth and division. The phenotype of cells during FtsZAT depletion (Movie 1) suggests that FtsZAT is also required, either directly or indirectly, for the proper termination of polar growth. If a subset of proteins involved in peptidoglycan biosynthesis or remodeling function during both polar elongation and septum formation, establishment of the FtsZAT ring could lead to disassembly of the polar elongation complex as the shared components are recruited to mid-cell. In addition, or alternatively, the FtsZAT-dependent establishment of new growth poles at mid-cell may trigger the cessation of growth from the old poles.
Table 1.
Quantitation of cell size and constriction of ftsZ mutants
| Average Cell Length† (μm +/− SD‡) | Average Cell Area† (μm2 +/− SD) | Average Constriction Rate‡ (nm/min +/− SD) | Relative Constriction Position§ +/− SD | ||
|---|---|---|---|---|---|
| WT | 2.31 +/−.50 | 1.66 +/−.35 | 6.82 +/−3.19 | .49 +/−.05 | |
| ∆ftsZ1 | 2.25 +/−.49 | 1.52 +/−.33 | 6.99 +/−3.58 | .46 +/−.05 | |
| ∆ftsZ3 | 2.24 +/−.47 | 1.44 +/−.30 | 6.77 +/−2.77 | .46 +/−.05 | |
| ∆ftsZ1 ∆ftsZ3 | 2.25 +/−.51 | 1.47 +/−.34 | 6.61 +/−3.75 | .46 +/−.04 | |
| ftsZAT depletion | -FtsZAT 0 h | 2.71 +/−.70 | 1.56 +/−.39 | 6.38 +/−2.81 | .49 +/−.07 |
| -FtsZAT 8 h | ND¶ | 2.95 +/−1.12 | ND | ND | |
| -FtsZAT 14 h | ND | 11.37 +/−4.69 | ND | ND | |
At least 100 cells were used to quantify the cell length and area for each strain.
At least 30 cells were used to quantify the constriction rates for each strain.
Relative constriction position for at least 40 cells is shown. A value of 0 corresponds to the new pole, 0.5 corresponds to mid-cell, and a value of 1 corresponds to the old pole.
ND – not determined.
Deletion of ftsZ1 and ftsZ3 does not change the FtsZAT depletion phenotype.
Since the ftsZ1 and ftsZ3 single deletions do not have an obvious impact on cell morphology, growth, or division, we constructed double and triple mutants to determine if there is an increasing effect when removing multiple ftsZ homologs. Double deletion of ftsZ1 and ftsZ3 does not cause a decrease in cell viability (Figure 2A, top panel), cell morphology (Table 1), or microcolony formation (Figure 2A, bottom panel). Furthermore, ΔftsZ1 ΔftsZ3 cells properly place constrictions and have an average constriction rate similar to WT (Table 1). Next, we introduced the ΔftsZ1, ΔftsZ3, and ΔftsZ1 ΔftsZ3 mutations into the ftsZAT depletion strain to determine if loss of multiple ftsZ homologs further aggravated the ftsZAT depletion phenotypes. The combination of the ftsZAT depletion strain with ΔftsZ1, ΔftsZ3, or ΔftsZ1 ΔftsZ3 mutations did not result in a further decrease in cell viability (Figure 2B, top panel), worsening of cell morphology (Figure 2B, bottom panel, Table 2), or accumulation of additional growth poles (Table 2) when compared to FtsZAT depletion alone. Together, these results suggest that the FtsZ1 and FtsZ3 homologs do not have a major impact on cell division under the conditions tested.
Figure 2. Removal of additional ftsZ homologs in the ftsZAT depletion strain does not cause a more severe phenotype.
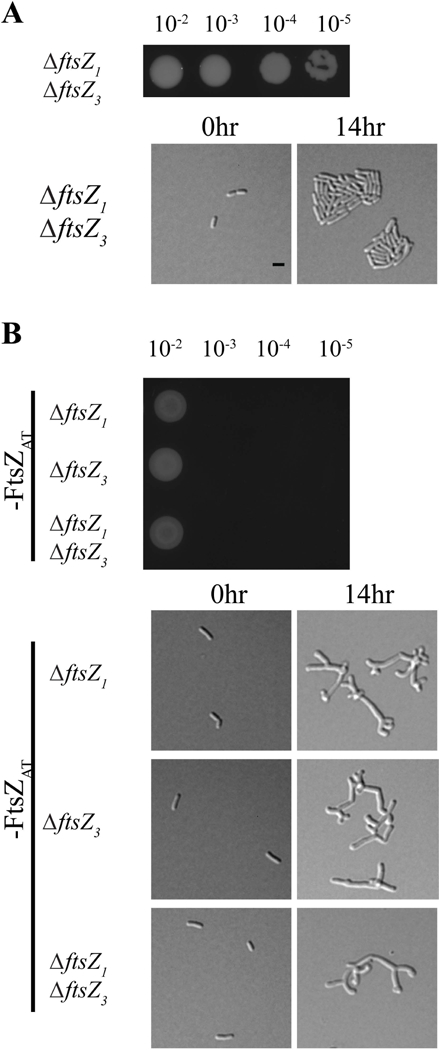
A) Cell viability (top) and morphology (bottom) of the double mutant ∆ftsZ1∆ftsZ3. B) Cell viability (top) and morphology (bottom) of ∆ftsZ1, ∆ftsZ3, or ∆ftsZ1∆ftsZ3 during FtsZAT depletion. All scale bars are set to 2 µm. Black bar denotes ftsZAT depletion strain background.
Table 2.
Quantitation of growth active and inactive poles and cell area
| Average Number Growth Inactive Poles† ±SD | Average Number Growth Active Poles† ±SD | Average Cell Area† (μm2 +/− SD) | ||
|---|---|---|---|---|
| ftsZAT depletion | -FtsZAT 14 h | 1.19 ±0.41 | 7.36 ±2.76 | 11.37 ±4.69 |
| -FtsZAT 14 h ΔftsZ1 | 1.14 ±0.38 | 7.22 ±2.77 | 10.89 ±3.25 | |
| -FtsZAT 14 h ΔftsZ3 | 1.16 ±0.42 | 7.10 ±2.89 | 11.12 ±5.61 | |
| -FtsZAT 14 h ΔftsZ1 ΔftsZ3 |
1.14 ±0.41 | 7.14 ±2.71 | 10.34 ± 4.27 | |
100 cells were used to quantify the number of growth active and inactive poles and cell area in each strain.
ftsZ gene duplications have occurred independently in several alphaproteobacterial lineages and in chloroplasts and some mitochondria (Vaughan et al., 2004). In most of the cases that have been studied, one FtsZ homolog plays a canonical role in cell or organelle division while the other plays a regulatory or specialized role. However, little is known about the roles of multiple FtsZs in certain alphaproteobacteria species. In both Rhizobium meliloti and Magnetospirillum gryphiswaldense, one of the FtsZs (containing a CTL and CTP similar to FtsZAT) is essential and the other (truncated after the GTPase domain similar to FtsZ1) is dispensable (Margolin & Long, 1994, Muller et al., 2014). In the case of M. gryphiswaldense, the truncated ftsZ is dispensable for division but important for biomineralization in this magnetotactic species under certain growth conditions. Similarly, it is possible that FtsZ1 or FtsZ3 may have important contributions to cell growth or division of A. tumefaciens in different environments such as in its plant-associated life-style.
FtsZ1 requires FtsZAT to localize to mid-cell and to polymerize in vitro.
Since FtsZ1 localizes to mid-cell (Figure 1B), we hypothesized that FtsZ1 may be a nonessential divisome component. To test this, we examined the localization of FtsZ1-sfGFP in both WT and the ftsZAT depletion strain (Figure 3). In WT and FtsZAT induced cells, FtsZ1-sfGFP does not localize in newborn cells but forms FtsZ-like rings at the future site of division in pre-divisional cells (Figure 3A, top and middle panel). This Z-like ring constricts to form a single focus in dividing cells. These observations suggest that FtsZ1 may be a divisome component despite the absence of a cell division phenotype in the ΔftsZ1 strain. To explore the possibility of interactions arising due to the loss of FtsZ1 and FtsZAT, we next visualized FtsZ1-sfGFP localization during the depletion of FtsZAT (Figure 3A, bottom panel). We pre-depleted FtsZAT for 4 h in liquid to avoid cell crowding caused by division events prior to sufficient FtsZAT depletion. Early during the depletion of FtsZAT, FtsZ1-sfGFP localizes in a FtsZ-like ring near mid-cell. However, as the FtsZAT depletion continues and branches begin to form, FtsZ1-sfGFP rings and foci progressively fade away, demonstrating that localization of FtsZ1-sfGFP to mid-cell likely requires the presence of FtsZAT.
Fgure 3. FtsZ1 requires FtsZAT to polymerize in vitro and to localize in cells.
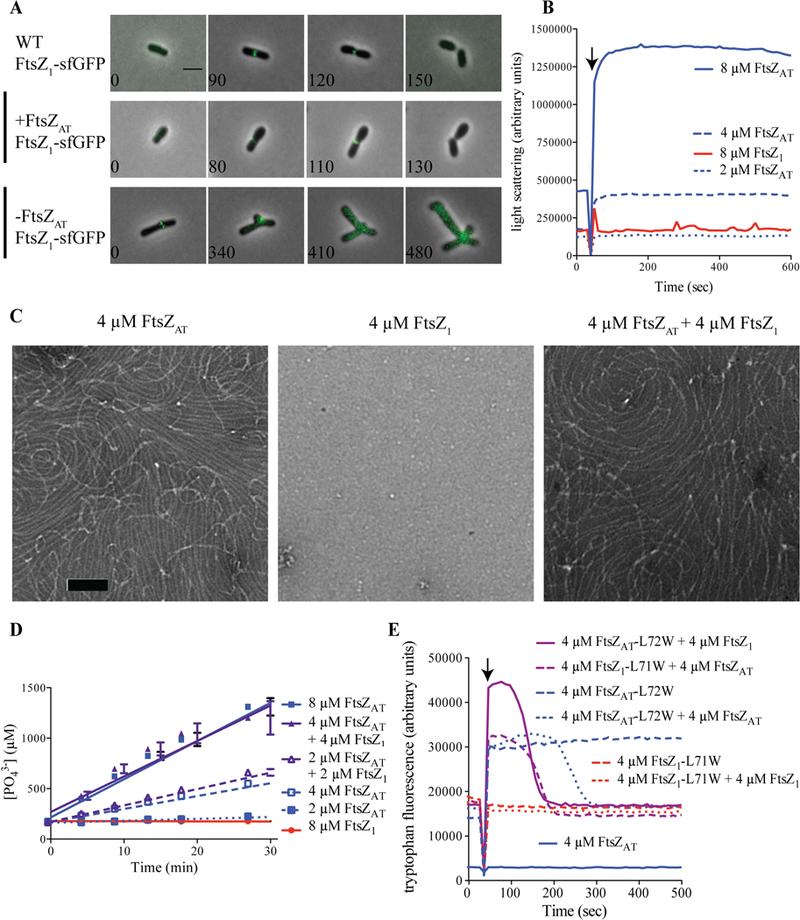
A) FtsZ1-sfGFP localization in WT, +FtsZAT and -FtsZAT. Scale bar is set to 2 µm. B) Light scattering over time for purified proteins at the indicated concentrations. GTP (2 mM) was added where indicated by the arrow to induce polymerization. Experiments were performed in triplicate and mean curves are shown. C) Negative stain TEM of the indicated proteins. Co-polymers of FtsZAT and FtsZ1 are indistinguishable from FtsZAT polymers. Scale bar is set to 100 nm. D) Inorganic phosphate concentration in solution over time in the presence of the indicated proteins and protein concentrations. Reactions were performed in triplicate and mean ± standard error is plotted. E) Tryptophan fluorescence over time for the indicated proteins. FtsZ1-L71W (red) shows no polymerization (Trp fluorescence) on its own, but can co-polymerize with added FtsZAT (purple). GTP (50 µM) was added where indicated by the arrow to induce polymerization. Experiments were performed in triplicate and representative curves are shown.
Since FtsZ1 is likely recruited to mid-cell by FtsZAT, we hypothesized that FtsZAT and FtsZ1 may form co-polymers. To first test the ability of FtsZAT and FtsZ1 to independently form polymers, each protein was purified (Supplemental Figure 1D) and subjected to polymerization studies. Right angle light scattering assays of wildtype FtsZAT revealed that this protein exhibits a GTP-dependent increase in light scattering at concentrations above 2 µm, consistent with its polymerization (Figure 3B, blue lines). Negative stain transmission electron microscopy (TEM) confirmed that FtsZAT forms gently curved protofilaments in the presence of GTP (Figure 3C, left panel) and it rapidly releases inorganic phosphate suggesting that GTP is hydrolyzed (Figure 3D, blue lines; 4.7 ± 0.2 GTP min−1 FtsZ−1 at 8 µM FtsZAT, n=3). Surprisingly, we did not observe polymerization of wildtype FtsZ1, even at high protein concentrations either in light scattering (Figure 3B, red line), TEM (Figure 3C, center panel), or GTP hydrolysis assays (Figure 3D, red line).
To determine if FtsZAT and FtsZ1 can form co-polymers, FtsZ1-L71W and FtsZAT-L72W were purified to enable monitoring of protein polymerization using tryptophan fluorescence (Figure 3E). The leucine to tryptophan mutation introduces a tryptophan on the surface of FtsZ that increases in fluorescence when it is buried in the subunit interface upon polymerization (Chen et al., 2005). While wildtype FtsZAT (with no tryptophan) does not change in fluorescence on addition of GTP (Figure 3F, solid blue line), FtsZAT-L72W fluorescence increases rapidly after GTP addition reflecting polymerization (Figure 3F, dashed blue line). When wildtype FtsZAT is added to FtsZAT-L72W, bringing the total FtsZ concentration to 8 µM, fluorescence again increases, but then drops back to baseline upon complete consumption of GTP by this high concentration of FtsZ (Figure 3F, dotted blue line). Conversely, on its own or combined with wildtype FtsZ1, FtsZ1-L71W maintains a constant tryptophan fluorescence level before and after addition of GTP, consistent with our conclusion that it does not polymerize on its own (Figure 3F, red lines). Remarkably, tryptophan fluorescence increases when FtsZ1-L71W and FtsZAT are mixed, indicating that the FtsZ1-L71W is incorporated into polymers in the presence of FtsZAT (Figure 3C, purple dashed line). When FtsZAT-L72W is mixed with FtsZ1, fluorescence increases above the level observed for FtsZAT-L72W alone and drops to baseline faster than FtsZAT-L72W on its own, again indicating co-polymerization. Finally, equimolar concentrations of FtsZAT alone or mixtures of FtsZAT and FtsZ1 exhibit similar rates of GTP hydrolysis (Figure 3D) and form qualitatively similar polymers by TEM (Figure 3C, right panel). Together, these observations indicate that FtsZ1 cannot polymerize independently, but that FtsZAT and FtsZ1 form co-polymers with similar structure and GTP hydrolysis rates as FtsZAT polymers.
Though multiple FtsZs are present in a number of bacterial and chloroplast lineages, their co-assembly properties have only begun to be characterized. In contrast to our observations, each copy of FtsZ in M. gryphiswaldense was able to independently polymerize in vitro, but they also appeared to directly interact, perhaps reflecting an ability to co-polymerize (Muller et al., 2014). Chloroplast FtsZs from Arabadopsis thaliana are also able to co-polymerize and, at least under some conditions, to independently polymerize (Olson et al., 2010). Conversely, one of the FtsZs from tobacco chloroplasts cannot polymerize on its own but promotes polymerization of its partner homolog (El-Kafafi el et al., 2005). Finally, the FtsZ pair from the chloroplasts of representative green and red algae co-polymerize into polymers with altered assembly dynamics from either homopolymer (TerBush et al., 2018). It is likely that in each of these cases, the assembly or co-assembly properties of the duplicated FtsZs have evolved to suit a niche regulatory function. We hypothesize the FtsZ1 from A. tumefaciens has low affinity for itself, but higher affinity for FtsZAT, limiting its homopolymerization but allowing for co-polymerization both in vitro and in cells. Since FtsZ1 cannot polymerize independently, FtsZAT must first polymerize at mid-cell after which FtsZ1 can be recruited by co-polymerization. The biological relevance of these biochemical and cell biological properties awaits further study.
FtsZAT depletion results in tip splitting events.
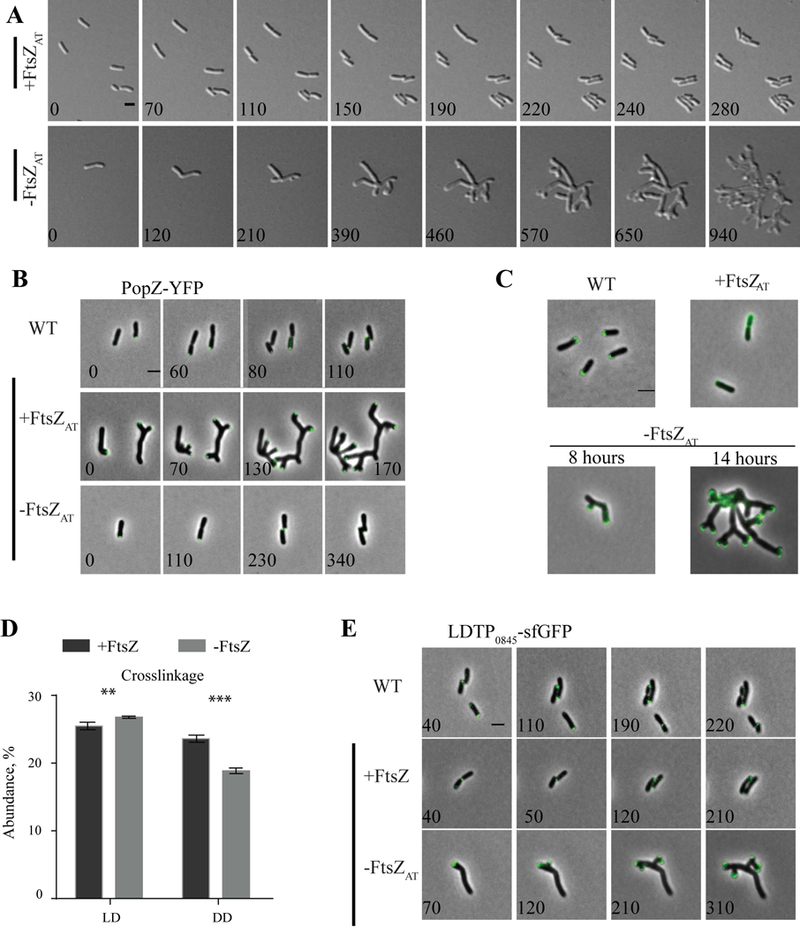
Once we identified FtsZAT as the primary homolog involved in cell division we next analyzed the growth phenotype during FtsZAT depletion more carefully. Compared to FtsZAT induced cells (Figure 4A, top), observation of cells during FtsZAT depletion by time-lapse microscope reveals remarkable changes in cell morphology (Figure 4A, bottom; Movie 1). Early during the depletion of FtsZAT, an ectopic pole forms near mid-cell. We hypothesize that this occurs due to the ability of the remaining FtsZAT to identify the mid-cell and recruit PG biosynthesis machinery to that site. Both the original growth pole and the ectopic pole are growth-active, resulting in the presence of multiple growth poles. These growth poles are unable to terminate cell elongation and ultimately most growth active poles are split, leading to the accumulation of many growth active poles (Figure 4A, bottom; Movie 1; Table 2) and the rapid increase in cell area (Table 1; Supplemental Figure 1C) until the cell lyses.
Figure 4. Characterization of continuous polar growth during FtsZAT depletion.
A) Timelapse microscopy of the ftsZAT depletion strain under inducing conditions (+FtsZAT, top panel) and depletion conditions (-FtsZAT, bottom panel). B) Timelapse microscopy showing PopZ-YFP localization WT, +FtsZAT and -FtsZAT. C) FDAA labeling of WT cells, and cells depleted of FtsZAT for 0 hours (+FtsZAT), 8 hours, and 14 hours. D) Abundance of total ld and dd crosslinkage in peptidoglycan isolated from ftsZAT depletion strain after induction (+FtsZAT, black bars) or depletion (-FtsZAT, grey bars) of FtsZAT for 14 hours. Data shown are the average abundance of each crosslinkage type and are taken from analysis of three independent biological samples. Statistical significance was calculated by t-tests and is indicated with an asterisk (P-value <0.05 (*), <0.005 (**), <0.001 (***)). E) Timelapse microscopy of LDTP0845-sfGFP in WT, +FtsZAT and -FtsZAT a. All scale bars are set to 2 µm.
The branched morphology observed during FtsZAT depletion is in stark contrast to FtsZ depletion observed in other organisms. In species like E. coli and B. subtilis, which utilize laterally localized peptidoglycan biosynthesis during elongation, depletion of FtsZ results in long, smooth filamentous cells. We hypothesize that the branching morphology of the A. tumefaciens FtsZAT depletion strain can be attributed to polar elongation. During the block in cell division, the growth pole continues to grow and presumably recruits additional peptidoglycan biosynthesis proteins. This could lead to an over-accumulation of proteins involved in cellular elongation causing the pole to split into two poles. A similar branching pattern has been characterized during typical growth of Streptomyces coelicolor (Richards et al., 2012). In this polar growing bacterium, the established complex of proteins contributing to elongation splits, leaving a small portion of the protein complex behind as growth continues. With time, the subpolar complex of proteins accumulates in size and eventually establishes a new growth pole. Although the polar growth molecular mechanisms are not conserved between A. tumefaciens and S. coelicolor, the fundamental principle of tip splitting as a consequence of polar growth appears to be shared.
PopZ-YFP accumulates at growth poles in the absence of FtsZAT.
In WT A. tumefaciens, deletion of popZ has been shown to cause ectopic poles and cells devoid of DNA, demonstrating a role in coordinating cell division with chromosome segregation (Howell et al., 2017a, Ehrle et al., 2017). We hypothesize that PopZ-dependent coordination of cell division likely involves FtsZ. In WT, PopZ-YFP localizes to the growing pole during elongation and is recruited to mid-cell just prior to cell separation (Figure 4B, top panel) (Ehrle et al., 2017, Howell et al., 2017a, Grangeon et al., 2015). In some freshly divided cells PopZ-YFP can be visualized at both the growing pole as well as the old pole resulting in an average of 1.12 PopZ-YFP labeled poles per cell (100 cells analyzed SD +/− .33). When FtsZAT is expressed in the ftsZAT depletion strain, PopZ-YFP has a similar localization pattern as in WT cells (Figure 4B, middle panel). When FtsZAT is depleted, PopZ-YFP stays at the growth poles and as tip splitting events lead to the production of new growth poles, PopZ-YFP appears to be split and retained at all growth active poles (Figure 4B, bottom panel). The accumulation of growing poles results in a population with an average 7.11 poles per cell containing PopZ-YFP (100 cells analyzed after 14 hours of FtsZAT depletion, SD +/− 2.65). These observations indicate that FtsZAT is required for recruitment of PopZ from the growth pole to mid-cell. Remarkably, both FtsZAT and FtsA are mislocalized in the absence of PopZ, leading to the establishment of asymmetric constriction sites and a broad range of cell lengths (Howell et al., 2017a). Together, these data suggest that the presence of both PopZ and FtsZ are important for proper positioning and functioning of the divisome.
Loss of septal PG synthesis results in altered total PG composition.
Since polar growth appears to continue in the absence of FtsZAT (Figure 4A, bottom panel), we used fluorescent-d-amino acids (FDAAs), to probe sites enriched in peptidoglycan synthesis (Kuru et al., 2012) during depletion of FtsZAT. In WT cells and conditions in which FtsZAT is expressed in the depletion strain, FDAAs localize at a single pole in elongating cells and at mid-cell in pre-divisional cells (Figure 4C, top) (Kuru et al., 2012). As FtsZAT is depleted, FDAAs are targeted strictly to the poles, confirming that polar peptidoglycan synthesis is responsible for the observed increase in cell biomass after 8 h and 14 h of depletion (Figure 4C, bottom).
Since cells depleted of FtsZAT fail to terminate polar growth and do not produce septal peptidoglycan, we hypothesized that the peptidoglycan composition may reveal chemical signatures of peptidoglycan derived from polar growth. Thus, we characterized the peptidoglycan composition of both WT cells and the ftsZAT depletion strain in both the presence and absence of IPTG using ultra-performance liquid chromatography (UPLC) (Alvarez et al., 2016). The major muropeptides found in WT A. tumefaciens PG and their quantification are shown in (Supplemental Figure 2A) and include monomeric (M), dimeric (D), and trimeric (T) muropeptides. The muropeptide composition and abundance is similar between WT cells, WT cells grown in the presence of IPTG, and the ftsZAT depletion strain grown in the presence of IPTG such that FtsZAT is expressed (Supplemental Figure 2B). These findings suggest that there are no major changes in PG composition due to IPTG and that the presence of IPTG leads to complementation in the ftsZAT depletion strain. In contrast, when the ftsZAT depletion strain is grown in the absence of IPTG for 14 h, marked changes in muropeptide composition are observed (Figure 4D, Supplemental Figure 2C-D). When FtsZAT is depleted, there is a significant increase in ld-crosslinkage as well as a decrease in total dd-crosslinkage (Figure 4D). These data suggest that the activity of ld-transpeptidases is increased and the activity of PBP-mediated dd-transpeptidases is decreased during FtsZAT depletion.
The A. tumefaciens genome contains 14 ld-transpeptidases, 7 of which are specific to Rhizobiales. The Rhizobiales-specific ld-transpeptidase encoded by Atu_0845 (referred to here as LDTP0845) has been shown to localize to the growing pole in WT cells and has been hypothesized to contribute to polar growth (Cameron et al., 2014). This localization pattern was confirmed in both WT and FtsZAT induced cells (Figure 4E). LDTP0845 is frequently found only at the growth pole, but also can exhibit a bipolar localization pattern. Quantification reveals that LDTP0845 localizes to an average of 1.18 poles per WT cell and 1.17 poles per cell expressing FtsZAT in the depletion strain (100 cells analyzed for each strain). We find that LDTP0845 localizes at 7.09 growth poles per cell following 14 hours of FtsZAT depletion (100 cells analyzed) (Figure 4E, bottom). This observation suggests that this LDTP0845 may contribute to changes in PG composition during FtsZAT depletion and supports a potential role for ld-transpeptidases in polar growth during elongation. Since there are 14 LDTPs in A. tumefaciens and thus far our preliminary data suggests that deleting any one of them does not cause a defect in polar growth, we suspect that redundant LDTPs likely contribute to polar growth.
FtsA is required for cell division but not initiation of growth from mid-cell.
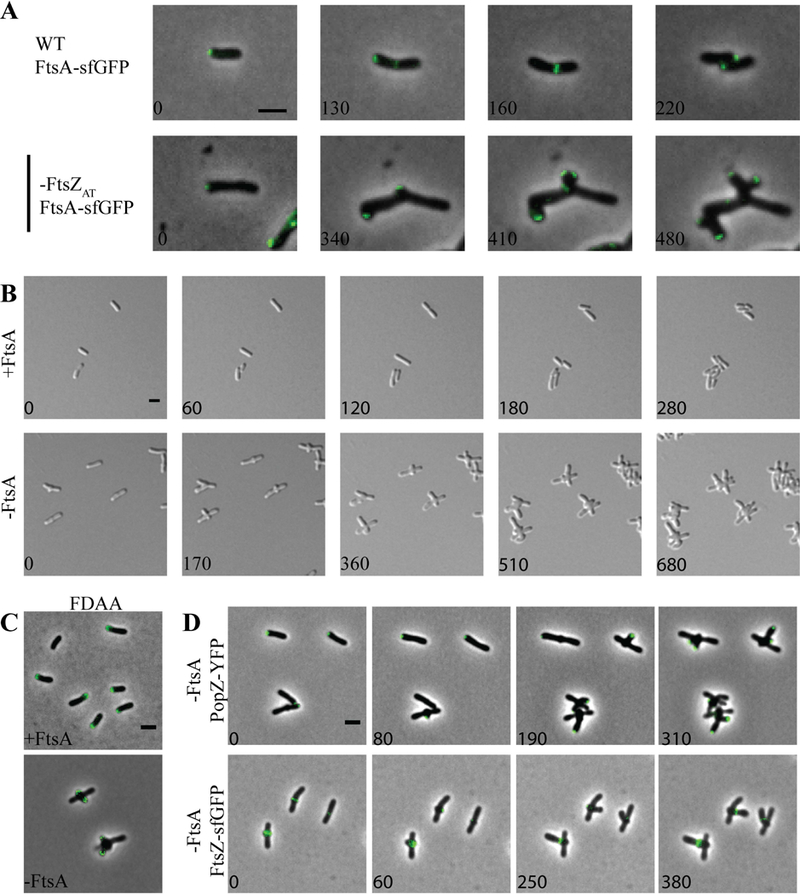
FtsA is an actin-like protein that associates with the membrane through an amphipathic helix and binds the FtsZ CTP to anchor FtsZ polymers to the membrane (Pichoff & Lutkenhaus, 2007, Szwedziak et al., 2012). In C. crescentus, recruitment of FtsA to mid-cell occurs well after the establishment of the FtsZ-ring and is dependent on the presence of FtsZ (Moll & Thanbichler, 2009, Goley et al., 2011). In A. tumefaciens, FtsA-sfGFP is retained at the growth pole prior to appearing at mid-cell just before cell division (Zupan et al., 2013, Cameron et al., 2014). Here, we confirm that FtsA-sfGFP is observed as a focus at the growth pole until transitioning to a ring-like structure at mid-cell (Figure 5A, top panel) At some timepoints, both a polar focus and a mid-cell ring of FtsA are observed (Figure 5A, top panel, 130 min). Eventually, the polar focus disappears as the FtsA-sfGFP ring becomes more intense just prior to cell division. During the depletion of FtsZAT, a focus of FtsA-sfGFP can be found at the growing pole, and at a newly formed ectopic pole near mid-cell (Figure 5A, bottom panel). FtsA-sfGFP remains associated with each growth pole, and as the poles undergo tip splitting events, each focus of FtsA-sfGFP is also split, resulting in the presence of FtsA-sfGFP in each of the 4 growth-active poles. These observations suggest that FtsZAT is required for proper localization of FtsA to mid-cell prior to cell division.
Figure 5. FtsA is not required for initiation of mid-cell growth.
A) FtsA-sfGFP localization in WT (top panel) and cells depleted of FtsZAT (bottom panel). B) Timelapse microscopy shows typical morphology when FtsA is induced and morphological changes in the absence of FtsA. C) FDAA labeling of the ftsA depletion strain after 8 hours of induction (+FtsA) or depletion (-FtsA). D) Timelapse microscopy of PopZ-YFP localization (top) and FtsZAT-GFP localization (bottom) during FtsA depletion. All scale bars are set to 2 µm.
Since FtsA tethers FtsZ to the membrane and enables divisome assembly (Vaughan et al., 2004, Szwedziak et al., 2012, Ma & Margolin, 1999) in E. coli, we expected that the depletion of FtsA would phenocopy the depletion of FtsZ. Although a saturating transposon mutagenesis screen indicated that ftsA is not essential for A. tumefaciens cell survival (Curtis & Brun, 2014), we were unable to construct a ∆ftsA mutant. Thus, we constructed a depletion strain in which expression of ftsA is controlled by Plac. Under conditions where FtsA is present in the ftsA depletion strain, cells maintain proper rod-shaped morphology, polar growth, and cell division occurs from constrictions formed near mid-cell (Figure 5B-C, top panels). In contrast, when FtsA is depleted, cells exhibit a marked change in morphology (Figure 5B, bottom panel; Movie 2). During the depletion of FtsA, rod-shaped cells initially elongate from a growth pole (Figure 5B, bottom panel, 0 min). Polar growth is terminated and growth is re-initiated from near mid-cell, typically resulting in the formation of two ectopic poles perpendicular to the original longitudinal axis of the cell (Figure 5B, bottom panel, 170 min). Cells depleted of FtsA continue multipolar growth (Figure 5B, bottom panel, 360 min), terminate growth from both poles and reinitiate growth from near mid-cell resulting in the formation of a new pair of ectopic growth poles (Figure 5B, bottom panel, 510 min). This pattern of multipolar growth, polar growth termination, and new branch formation is continued until cells eventually bulge at the mid-cell and lyse. Overall these observations indicate that the phenotypes caused by FtsZAT and FtsA depletion are distinct from one another and suggest that only FtsZ is required for initiation of PG biosynthesis at mid-cell and the cessation of polar growth.
To confirm that polar growth occurs and is terminated during FtsA depletion, cells were labeled with FDAAs (Figure 5C, bottom panel). Indeed, FDAAs label the tips of two poles, which are emerging from near mid-cell consistent with the re-initiation of polar growth. To further confirm that polar growth is terminated during FtsA depletion, we observed the localization of PopZ-YFP as FtsA is depleted (Figure 5D, top panel). PopZ marks the growth poles (Grangeon et al., 2015) and becomes trapped at growth poles during depletion of FtsZ (Figure 4B). During FtsA depletion, PopZ-YFP is initially present at the growth pole (Figure 5D, top panel, 0 min). Next, PopZ-YFP disappears from the growth poles and reappears near mid-cell (Figure 5D, top panel, 80 min) indicating that growth is redirected to mid-cell. Throughout the FtsA depletion, PopZ-YFP continues to disappear from growth poles and reappears at the tips of newly emerging growth poles. Overall, these observations clearly indicate that FtsA is not necessary for termination of polar growth and initiation of growth from mid-cell; however, FtsA has an essential function at a later stage of cell division since the cells fail to divide and are prone to lysis.
The ability of cells to target growth to near mid-cell during FtsA depletion suggests that FtsZ-rings may form, enabling the termination of polar growth. Indeed, FtsZAT-sfGFP-rings form near mid-cell early during FtsA depletion (Figure 5D, bottom panel). FtsZAT-sfGFP is briefly retained at new growth poles before reappearing to mark the site where a new growth pole will emerge. These observations are consistent with the finding the FtsA is retained at the growth pole longer than FtsZ (Cameron et al., 2015, Zupan et al., 2013), and suggest that FtsA arrives at mid-cell after Z-ring assembly and the initiation of FtsZ-dependent cell wall biogenesis. The results observed here in A. tumefaciens are consistent with the observation that FtsA arrives to mid-cell after FtsZ and the onset of mid-cell cell wall biogenesis in C. crescentus (Moll & Thanbichler, 2009, Goley et al., 2011). In both A. tumefaciens and C. crescentus, the late arrival of FtsA to the divisome suggests that other proteins contribute to proper tethering of FtsZ to the membrane. In C. crescentus, the FtsZ-binding protein, FzlC, functions as a membrane anchor early during the establishment of the divisome (Goley et al., 2010, Meier et al., 2016). A homolog of FzlC is readily found in the A. tumefaciens genome (Atu2824) and may contribute to the ability of FtsZ-rings to form in the absence of FtsA.
Depletion of the downstream divisome component FtsW phenocopies depletion of FtsA.
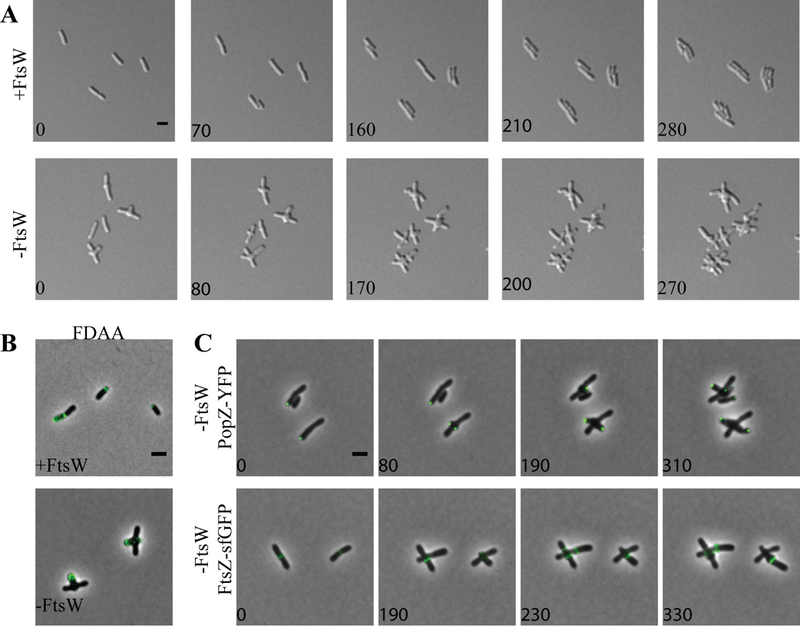
Having observed a distinct effect on cell morphology in the absence of ftsA, we wondered if the phenotype observed during ftsA depletion could be recapitulated in the absence of another late-arriving divisome protein. To test this hypothesis, we constructed a depletion strain of FtsW, which is recruited to mid-cell after FtsA in both E. coli and C. crescentus divisome assembly models (Goley et al., 2011, Du & Lutkenhaus, 2017). Depletion of FtsW results in a phenotype which is strikingly similar to the depletion of FtsA (Figure 6). When FtsW is induced normal growth is observed (Figure 6A, top panel). During FtsW depletion, polar growth is terminated, resulting in the establishment of growth-active poles from near mid-cell (Figure 6A, bottom panel; Movie 3). Multiple rounds of termination of polar growth followed by reinitiation of growth from near mid-cell occur until the mid-cell bulges and the cells ultimately lyse (Figure 6A, bottom panel). Labeling of growth active poles with FDAAs (Figure 6B) or by tracking PopZ-YFP localization (Figure 6C, top panel) confirmed that new branches which emerge from mid-cell are formed by polar growth. Finally, we confirmed that FtsZ-rings form during the depletion of FtsW and the presence of an FtsZAT-sfGFP-ring typically marks the site where an ectopic growth pole will form (Figure 6C, bottom panel). Together, these observations suggest that FtsZ-rings are formed in the absence of FtsW, enabling the initiation of cell wall biogenesis. Given that FtsW likely drives septal PG biosynthesis, (Fraipont et al., 2011) these findings indicate that the cell wall biogenesis that occurs during depletion of FtsA or FtsW may require the elongation machinery, which typically functions in polar growth. Since the elongation machinery for A. tumefaciens remains to be identified, it is possible that there is considerable overlap between the machineries that contribute to polar and septal PG biosynthesis.
Figure 6. FtsW is not required for initiation of mid-cell growth.
A) Timelapse microscopy of the ftsW depletion strain when induced (+FtsW, top) or depleted (-FtsWm bottom). B) FDAA labeling of the ftsW depletion strain after 8 hours of induction (+FtsW) or depletion (-FtsW). C) Timelapse microscopy of PopZ-YFP localization (top) and FtsZAT-GFP localization (bottom) during FtsW depletion. All scale bars are set to 2 µm.
Concluding Remarks
While many questions remain unanswered about the regulation of cell wall biogenesis in A. tumefaciens, our work sheds light on the transition from polar growth to mid-cell growth. We find that FtsZAT, FtsA, and FtsW are required for constriction and cell separation, but FtsZAT is also required to terminate polar growth and initiate mid-cell peptidoglycan synthesis. How might the formation of an FtsZAT-ring at mid-cell cause the termination of polar growth? We find that PopZ, and LDTP0845 become trapped at the growth poles during FtsZ depletion (Figure 5). It is possible that one or more of these proteins contributes to both polar peptidoglycan biosynthesis and mid-cell peptidoglycan synthesis and that the FtsZ-dependent targeting of these proteins (and likely others) to mid-cell triggers the termination of polar growth. While the mid-cell localization of PopZ is dependent on the presence of FtsZAT (Figure 4), FtsZAT-ring stability and placement are impacted by the absence of PopZ (Howell et al., 2017a). Furthermore, deletion of popZ impairs termination of polar growth and results in cell division defects (Howell et al., 2017a, Ehrle et al., 2017, Grangeon et al., 2017). The apparent co-dependence of FtsZ and PopZ for localization may suggest that these proteins function together during the early stages of cell division, particularly the termination of polar growth and onset of mid-cell PG biosynthesis.
Overall, our results are consistent with a general model, which is highly conserved in bacteria, in which the establishment of a FtsZ-ring leads to the recruitment of many other cell division proteins to mid-cell (Du & Lutkenhaus, 2017), though many mechanistic questions remain. How is FtsZAT targeted to mid-cell? A variety of mechanisms that contribute to the proper placement of FtsZ at mid-cell have been described (for review see (Rowlett & Margolin, 2015, Monahan et al., 2014)). The most well studied mechanisms of FtsZ positioning include negative regulation by the Min system and nucleoid occlusion. While genes encoding components of the Min system are readily identifiable in the A. tumefaciens genome, the deletion of minCDE has a minimal impact on placement of constriction sites and cell division efficiency (Flores et al., 2018). Furthermore, FtsZAT-GFP rings form over DNA prior to nucleoid separation in A. tumefaciens. These observations indicate that additional regulatory mechanisms must contribute to proper division site selection in A. tumefaciens. Following the appearance of FtsZ at mid-cell, how is the FtsZAT-ring stabilized? In E. coli, the FtsZ-ring is stabilized by interactions with FtsA and ZipA, which tether FtsZ filaments to the membrane (Ma & Margolin, 1999, Mosyak et al., 2000, Haney et al., 2001, Szwedziak et al., 2012). In A. tumefaciens, FtsZAT appears at mid-cell well before FtsA (Cameron et al., 2014) and we observe that FtsZAT rings form even when FtsA is depleted (Figure 5D, bottom panel). Furthermore, the position of FtsZAT-GFP rings marks the site of ectopic pole formation. These observations suggest the FtsZAT is stabilized, at least early during cell division by other proteins. While there are no obvious ZipA homologs encoded in the A. tumefaciens genome, a homolog of FzlC, which functions to stabilize FtsZ in C. crescentus (Goley et al., 2010, Meier et al., 2016), is encoded in the genome.
The observation that FtsZ is necessary for the initiation of mid-cell PG biosynthesis suggests that FtsZ is necessary for recruitment of PG biosynthesis enzymes to mid-cell. Septal PG biosynthesis is likely mediated by FtsW, a putative PG glycosyltransferase (Cho et al., 2016, Meeske et al., 2016, Emami et al., 2017), and PBP3 (FtsI), a PG dd-transpeptidase (Botta & Park, 1981). In A. tumefaciens, depletion of FtsW does not cause a complete block of PG synthesis at mid-cell (Figure 5). This observation suggests that mid-cell PG biosynthesis is mediated by other cell wall biogenesis enzymes while the activity of FtsW contributes to later stages of cell division, consistent with the inability of cells to form constrictions and separate in the absence of FtsW. These observations may indicate that the initial PG biosynthesis at mid-cell comprises the final stage of cell elongation, consistent with descriptions of FtsZ-dependent mid-cell elongation in C. crescentus (Aaron et al., 2007). The observation that growth-active, ectopic poles emerge from near mid-cell during FtsW depletion (Figure 5B) provides evidence in support of this possibility. Thus, FtsZ-dependent PG biosynthesis may contribute to both elongation and cell division in A. tumefaciens. For a polar growing bacterium, it is tempting to speculate that the retention of PG biosynthesis machinery dedicated to elongation at the site of cell division may prime the newly formed poles to become growth active following cell separation.
Experimental Procedures
Bacterial strains and culture conditions.
All bacterial strains and plasmids used are listed in Table S1. A. tumefaciens strains were grown in ATGN minimal medium with .5% glucose (Morton & Fuqua, 2012a) at 28°C. E. coli strains were grown in Luria-Bertani medium at 37°C. When indicated, kanamycin (KM) was used at 300 μg ml−1 for A. tumefaciens, 50 μg ml−1 for E. coli DH5α, and 25 μg ml−1 for E. coli S17–1 λ pir. Gentamicin was used when indicated at 200 μg ml−1 for A. tumefaciens and 20 μg ml−1 for E. coli DH5α. IPTG was added at a concentration of 1 mM when indicated. Cumate was added at a concentration of 0.1 mM when indicated.
Construction of expression plasmids and strains.
All strains and plasmids used are listed in Table S1, while primers used are listed in Table S2. For amplification of target genes, primer names indicate the primer orientation and added restriction sites. To construct expression vectors containing ftsZAT-sfgfp, ftsZ1-sfgfp, ftsZ3-sfgfp, and ldtp0845-sfgfp the respective coding sequence was amplified from purified C58 genomic DNA using primers indicated in Table S4.2. The amplicons were digested overnight and ligated into cut pSRKKM-Plac-sfgfp using NEB T4 DNA ligase at 4°C overnight. The newly formed sfgfp fusion of each gene was excised from the plasmid by overnight digestion with NdeI and NheI. Fragments containing ftsZAT-sfgfp, ftsZ1-sfgfp, ftsZ3-sfgfp, and ldtp0845-sfgfp were then ligated into cut pRV-MCS2 to give constitutive expression vectors containing the fusions. To construct the popZ-yfp expression vector, popZ along with the upstream promoter sequence were amplified from purified C58 genomic DNA, digested and ligated into pMR10 containing YFP.
Construction of deletion/depletion plasmids and strains.
Vectors for gene deletion by allelic exchange were constructed using recommended methods for A. tumefaciens (Morton & Fuqua, 2012b). Briefly, 500 bp fragments upstream and downstream of the target gene were amplified using primer pairs P1/P2 and P3/P4 respectively. Amplicons were spliced by overhang extension (SOE) using primer pair P1/P4. The amplicon was digested and ligated into pNTPS139. The deletion plasmids were introduced into A. tumefaciens by mating using an E. coli S17 conjugation strain to create kanamycin (KM) resistant, sucrose sensitive primary exconjugants. Primary exconjugants were grown overnight in media with no selection. Secondary recombinants were screened by patching for sucrose resistance and KM sensitivity. Colony PCR with primers P5/P6 for the respective gene target was used to confirm deletion. PCR products from P5/P6 primer sets were sequenced to further confirm deletions.
For depletion strains, target genes (ftsZAT, ftsA, and ftsW) were amplified, digested and ligated into either pUC18-mini-Tn7T-GM-Plac or pUC18-mini-Tn7T-GM-Plac. The mini-Tn7 vectors, along with the pTNS3 helper plasmid, were introduced into C58∆tetRA::a-attTn7 as described previously (Figueroa-Cuilan et al., 2016). Transformants were selected for gentamicin resistance and insertion of the target gene into the a-att site was verified by colony PCR using the tet forward and Tn7R109 primer. PCR products were sequenced to confirm insertion of the correct gene. Next, the target gene was deleted from the native locus as described above in the presence of 1 mM IPTG to drive expression of the target gene from the engineered site.
Construction of plasmids for protein expression and purification.
To construct pET21a FtsZAT, ftsZAT was amplified from C58 genomic DNA with FtsZAT For NdeI and FtsZAT Rev EcoRI, digested with NdeI and EcoRI, and ligated into similarly digested pET21a. To construct pTB146 FtsZ1, ftsZ1 was amplified from C58 genomic DNA with FtsZ1 For SapI and FtsZ1 Rev BamHI, digested with SapI and BamHI, and ligated into similarly digested pTB146. Ligation products were transformed into NEB Turbo (New England Biolabs) and selected for ampicillin resistance. Insertions were verified by colony PCR and Sanger sequencing. Primers FtsZAT-L72W and FtsZ1-L71W were used to mutagenize pET21a FtsZAT and pTB146 FtsZ1, respectively, using the Quikchange Multi Lightning Mutagenesis Kit (Agilent) and following the manufacturer’s protocol to generate pET21a FtsZAT-L72W and pTB146 FtsZ1-L71W. Mutations in the targeted sites were verified by Sanger sequencing.
DIC and phase contrast microscopy.
Exponentially growing cells (OD600 = ~0.6) were spotted on 1% agarose ATGN pads as previously described (Howell et al., 2017b). Microscopy was performed with an inverted Nikon Eclipse TiE with a QImaging Rolera em-c2 1K EMCCD camera and Nikon Elements Imaging Software. For time-lapse microscopy, images were collected every ten minutes, unless otherwise stated.
Fluorescence microscopy.
Plasmid encoded FtsZAT-sfGFP, FtsZ1-sfGFP, FtsZ3-sfGFP, and LDTP0845-sfGFP fusions were expressed from the Pvan promoter, which provides constitutive low levels of expression (Figure 6- Figure Supplement 1C). To construct demographs of various FtsZ-sfGFP localization, at least 225 cells were imaged for each strain. A GFP channel profile was taken along the medial axis for each cell imaged. These medial axis profiles were then aligned by cell length using MicrobeJ software (Ducret et al., 2016).
Plasmid encoded FtsA-sfGFP and PopZ-YFP fusions were expressed from the native promoters. Cells containing plasmids with fluorescent protein fusions were grown to exponential phase before imaging on agarose pads.
To visualize sites of active peptidoglycan synthesis 1 ml of exponentially growing cells was labeled with 5 μl of 5mM fluorescent D-amino acid (FDAA), HCC amino-D-alanine (HADA) for 1 minute and washed three times to remove residual label from the supernatant, as previously described (Kuru et al., 2012, Howell et al., 2017b).
Cell viability and growth curve assays.
For cell viability spot assays, cells were grown with IPTG to exponential phase before washing 3 times to remove inducer. Cells were then diluted to OD600 = 0.1 and serially diluted in ATGN (no inducer). 3 µl of each dilution was spotted onto ATGN and incubated at 28°C for 3 days before imaging. When appropriate ATGN plates contained KM 300 μg ml−1, and IPTG 1mM as indicated in figure legends. For growth curve analysis, exponentially growing cultures were diluted to OD600 = .05 in 200 µl of ATGN in 96-well plates. Plates were shaken for 1 minute before OD600 readings, which were taken every 10 minutes.
Cell morphology and constriction rate analysis.
Exponentially growing cells were imaged using phase contrast microscopy as described above. Cell length, area, and constrictions were detected using MicrobeJ software (Ducret et al., 2016). To calculate constriction rates, cells with detectable constrictions were tracked using time-lapse microscopy. The width of the cell constriction was measured at an initial time-point and the measurement was repeated after 10 minutes. The difference in constriction width was divided by the 10-minute time interval to give a constriction rate.
To quantify the number of growth active and inactive poles, cells were grown to exponential phase with IPTG and washed 3 times before adding fresh media with or without IPTG. Cells were cultured under normal growth conditions for 14 hours before labelling active growth poles with FDAAs as described above. FDAA labelled poles were then quantified using MicrobeJ software.
Western blot analysis.
For western blot analysis of FtsZ depletion, the ftsZ depletion strain was grown in 40 ml ATGN with 1 mM IPTG to exponential phase. 2 ml of culture was collected prior to depletion (time 0) by centrifugation at 10,000 x g for 3 minutes. The remaining culture was collected by centrifugation at 3500 x g for 10 minutes, and supernatants were discarded. Cells were washed in sterile water and pelleted again. To deplete FtsZ, the pellet was resuspended in fresh ATGN without IPTG and grown under standard culturing conditions. 2-ml samples were collected by centrifugation after 30, 45, 60, 120, and 240 minutes of depletion. OD600 was taken for each sample prior to centrifugation so that samples could be normalized to an OD600 equivalent to 0.68. The cell pellets were incubated with 100 μl of a master mix containing 1 ml of BugBuster protein extraction reagent (Novagen) and supplemented with 1 EDTA-free protease inhibitor cocktail (Sigma), 10 μl of lysonase (Novagen), 2,500 U ml−1 DNase I (Thermo Scientific), and 1 mM dithiothreitol (DTT) (Thermo Scientific) for 25 minutes with shaking at room temperature to lyse the cell pellets. The whole-cell lysates were clarified by centrifugation at 10,000 rpm for 15 min. A final concentration of 1 X Laemmli buffer was added to the cleared cell lysates. Samples were boiled at 100°C for 5 min prior to loading on a 4–15% Mini-PROTEAN TGX Precast Gel (Bio-Rad). The separated proteins were electroblotted onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad) and blocked overnight in 5% nonfat dry milk powder solubilized in 1% TBST (Tris-buffered saline [TBS], 1% Tween 20). The blocked PVDF membranes were probed with Escherichia coli anti-FtsZ serum at a final concentration of 1/3000 ((Ward & Lutkenhaus, 1985), gift from Joe Lutkenhaus) for 1.5 h in 5% milk-TBST, followed by incubation with anti-rabbit (1:5000) HRP (Pierce 31460) secondary antibody for 1 h in 5% milk-TBST. The secondary antibody was detected using the ECL Plus HRP substrate (Thermo Scientific Pierce).
Protein expression and purification.
FtsZAT and FtsZAT-L72W were expressed and purified in untagged form. Each was produced from a pET21 expression vector (pEG1555 – FtsZAT, pEG1556 - FtsZAT-L72W,) in Escherichia coli Rosetta(DE3)pLysS induced at 37°C for 4 h with 0.5 mM IPTG after OD reached 0.8 to 1.0 OD at 600 nm. Cells were harvested by centrifugation at 6000 x g and resuspended in 30 mL FtsZ QA buffer (50 mM Tris-HCl pH 8, 50 mM KCl, 0.1 mM EDTA, 10% glycerol) per liter of culture. Resuspensions were snap frozen in liquid nitrogen and stored at −80°C until purification. To purify, resuspensions were thawed quickly and cells were lysed by incubation with 1 mg ml−1 lysozyme, 2.5 mM MgCl2, DNAse I, 2 mM PMSF, and a complete mini EDTA-free protease inhibitor tablet (Roche) for 45 min to 1 h at room temperature followed by sonication. Lysates were cleared by centrifugation at 15000 x g for 30 min at 4°C and filtered through a 0.45 µm filter before anion exchange chromatography (HiTrap Q HP 5 mL, GE Life Sciences). Protein was eluted with a linear KCl gradient (FtsZ QA buffer with 50 to 500 mM KCl) and fractions containing FtsZ were verified by SDS-PAGE, pooled, and subjected to ammonium sulfate precipitation. Precipitates (at 17–20% ammonium sulfate saturation depending on the variant) were verified by SDS-PAGE, resuspended in HEK50G (50 mM HEPES-KOH pH 7.2, 0.1 mM EDTA, 50 mM KCL, 10% glycerol, 1 mM β-mercaptoethanol), and further purified by gel filtration (Superdex 200 10/300 GL, GE Life Sciences). Peak fractions were pooled, snap frozen in liquid nitrogen, and stored at −80°C.
FtsZ1 and FtsZ1-L71W were produced as His6-SUMO fusions and cleaved to yield untagged, scarless proteins. Each was produced from a pTB146 expression vector (pEG1535 - FtsZ1, pEG1542 - FtsZ1-L71W) in E. coli Rosetta (DE3)pLysS as described above. Cells were harvested by centrifugation as above, resuspended in HK300G (50 mM HEPES-KOH pH7.2, 300 mM KCl, 10% glycerol) with 20 mM imidazole, snap frozen in liquid nitrogen, and stored at −80°C until purification. To purify, resuspensions were thawed quickly and cells were lysed by incubation with 1 mg ml−1 lysozyme, 2.5 mM MgCl2, and DNase I for 45 min at room temperature followed by sonication. Lysate was cleared and filtered as described above. Protein was isolated by Ni2+ affinity chromatography (HisTrap FF 1 mL, GE Life Sciences) and eluted in HK300G with 300 mM imidazole. Fractions containing His6-SUMO fusions were verified by SDS-PAGE, combined with Ulp1 Sumo protease at a 1:100 (protease:FtsZ) molar ratio, and cleaved by incubation at 30°C for 3.5 h. Cleaved FtsZ1 or FtsZ1L71W was purified away from His6-SUMO by gel filtration (Superdex 200 10/300 GL, GE Life Sciences) in HEK50G. Peak fractions were pooled, snap frozen in liquid nitrogen, and stored at −80°C.
Polymerization kinetics assays.
A Fluoromax-3 spectrofluorometer (Jobin Yvon, Inc) was used to monitor FtsZ polymerization by right-angle light scattering and tryptophan fluorescence. FtsZ1 and/or FtsZAT (wild-type or L71W/L72W mutants, as indicated in figures and text) was polymerized in HEK50 (50 mM HEPES-KOH pH 7.2, 50 mM KCl, 0.1 mM EDTA) with 2.5 mM MgCl2. 2 mM GTP was used to induce polymerization for light scattering and 50 µM GTP was used to induce polymerization for tryptophan fluorescence (GTP is fluorescent at the wavelengths used, so low concentrations must be used). GTP was added after baseline light scatter or fluorescence was established. For light scattering, samples were excited at 350 nm and scatter was detected at 350 nm with slits set to 2 nm. For tryptophan fluorescence, samples were excited at 295 nm and emission was detected at 344 nm, with 2 nm slits.
GTPase assay.
FtsZ1 and/or FtsZAT was polymerized in HEK50 with 2.5 mM MgCl2 and 2 mM GTP. GTP was added at time 0. Reaction was stopped at 5, 10, 15, 20, and 30 minutes with quench buffer (50 mM HEPES-KOH pH 7.2, 21.3 mM EDTA, 50 mM KCl). Inorganic phosphate in solution (liberated by GTP hydrolysis) over time was measured using SensoLyte MG Phosphate Assay Kit Colorimetric (AnaSpec, Inc, Fremont, California).
Negative stain transmission electron microscopy (TEM).
FtsZ1 and/or FtsZAT were polymerized in HEK50 with 2.5 mM MgCl2 and 2 mM GTP. After a 15-minute incubation at room temperature, samples were applied to carbon-coated glow-discharged grids with 0.75% uranyl formate staining as previously described (Lariviere et al., 2018, Sundararajan et al., 2015). TEM samples were imaged using a Philips/FEI BioTwin CM120 TEM equipped with an AMT XR80 8 megapixel CCD camera (AMT Imaging, USA).
Peptidoglycan composition analysis
Six cultures of WT and ftsZ depletion cells were grown in 10 ml of ATGN with IPTG to exponential phase. The 10 ml cell cultures were added to 40 ml of fresh media. The 50 ml cultures were grown to exponential phase and pelleted by centrifugation at 4000 x g for 10 minutes. Cell pellets were washed three times with ATGN by centrifugation and resuspension to remove IPTG. After the final wash 3 cell pellets were resuspended in 50 ml ATGN and the remaining 3 pellets were resuspended in 50 ml ATGN with 1 mM IPTG. Each culture was grown for 14 h. The optical densities of the cells were monitored to ensure the optical density of the cultures never went above OD600 = 0.7 to avoid changes to peptidoglycan content due to stationary phase. If necessary, fresh medium was added to dilute the cultures to maintain exponential growth. After 14 h of growth, 50 ml of the exponential cultures were collected and pelleted by centrifugation at 4000 x g for 20 minutes. Cell pellets were resuspended in 1mL of ATGN and 2 mL of 6% SDS and stirred with magnets while boiling for 4 h. After 4 h, samples were removed from heat but continued to stir overnight. Samples were then shipped to Dr. Felipe Cava’s laboratory for purification and analysis.
Upon arrival, cells were boiled and simultaneously stirred by magnets for 2 h. After 2 h, boiling was stopped and samples were stirred overnight. Peptidoglycan was pelleted by centrifugation for 13 min at 60000 rpm (TLA100.3 Beckman rotor, Optima Max-TL ultracentrifuge; Beckman), and the pellets were washed 3 to 4 times by repeated cycles of centrifugation and resuspension in water. The pellet from the final wash was resuspended in 50 µl of 50 mM sodium phosphate buffer, pH 4.9, and digested overnight with 100 µg ml−1 of muramidase at 37°C. Muramidase digestion was stopped by boiling for 4 min. Coagulated protein was removed by centrifugation for 15 min at 15000 rpm in a desktop microcentrifuge. The muropeptides were mixed with 15 µl 0.5 M sodium borate and subjected to reduction of muramic acid residues into muramitol by sodium borohydride (10 mg ml−1 final concentration, 20 min at room temperature) treatment. Samples were adjusted to pH 3 to 4 with orthophosphoric acid and filtered (0.2-µm filters).
Muropeptides were analyzed on a Waters UPLC system equipped with an ACQUITY UPLC BEH C18 Column, 130 Å, 1.7 μm, 2.1 mm × 150 mm (Waters) and a dual wavelength absorbance detector. Elution of muropeptides was detected at 204 nm. Muropeptides were separated at 45°C using a linear gradient from buffer A [formic acid 0.1% (v/v)] to buffer B [formic acid 0.1% (v/v), acetonitrile 20% (v/v)] in a 12 min run with a 0.250 ml min−1 flow. Peptidoglycan compositional analysis on triplicate samples was completed on two separate occasions.
Supplementary Material
Acknowledgements
We thank Yves Brun from providing plasmids, Joe Lutkenhaus for providing Escherichia coli anti-FtsZ serum, and members of the Brown lab for helpful discussions and critical reading of this manuscript. PB and MH were supported by the National Science Foundation, IOS1557806. This work was funded in part by the National Institutes of Health through R01GM108640 (EDG) and T32GM007445 (training support of PJL). FC and AA receive funding support from Laboratory for Molecular Infection Medicine Sweden, Knut and Alice Wallenberg Foundation, Kempe and the Swedish Research Council. AA is supported by a MIMS/VR PhD position.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, and Jacobs-Wagner C (2007) The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 64: 938–952. [DOI] [PubMed] [Google Scholar]
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, and den Blaauwen T (2005) Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol 55: 1631–1645. [DOI] [PubMed] [Google Scholar]
- Alvarez L, Hernandez SB, de Pedro MA, and Cava F (2016) Ultra-sensitive, high-resolution liquid chromatography methods for the high-throughput quantitative analysis of bacterial cell wall chemistry and structure. Methods in molecular biology (Clifton, N.J.) 1440: 11–27. [DOI] [PubMed] [Google Scholar]
- Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, and De Bolle X (2002) Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol 43: 945–960. [DOI] [PubMed] [Google Scholar]
- Bi EF, and Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164. [DOI] [PubMed] [Google Scholar]
- Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, Brun YV, and Garner EC (2017) Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science (New York, N.Y.) 355: 739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta GA, and Park JT (1981) Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. Journal of bacteriology 145: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJB, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, and Brun YV (2012) Polar growth in the Alphaproteobacterial order Rhizobiales. Proceedings of the National Academy of Sciences of the United States of America 109: 1697–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron TA, Anderson-Furgeson J, Zupan JR, Zik JJ, and Zambryski PC (2014) Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. mBio 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron TA, Zupan JR, and Zambryski PC (2015) The essential features and modes of bacterial polar growth. Trends in Microbiology 23: 347–353. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bjornson K, Redick SD, and Erickson HP (2005) A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophysical journal 88: 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Sibley CD, Zaheer R, and Finan TM (2007) A Sinorhizobium meliloti minE mutant has an altered morphology and exhibits defects in legume symbiosis. Microbiology (Reading, England) 153: 375–387. [DOI] [PubMed] [Google Scholar]
- Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PD, Suh H, Marto JA, Garner EC, and Bernhardt TG (2016) Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nature microbiology: 16172. [DOI] [PMC free article] [PubMed]
- Curtis PD, and Brun YV (2014) Identification of essential alphaproteobacterial genes reveals operational variability in conserved developmental and cell cycle systems. Mol Microbiol 93: 713–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Crossley R, and Rothfield L (1992) The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359: 254–256. [DOI] [PubMed] [Google Scholar]
- den Blaauwen T, Andreu JM, and Monasterio O (2014) Bacterial cell division proteins as antibiotic targets. Bioorganic chemistry 55: 27–38. [DOI] [PubMed] [Google Scholar]
- Du S, and Lutkenhaus J (2017) Assembly and activation of the Escherichia coli divisome. Mol Microbiol 105: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A, Quardokus EM, and Brun YV (2016) MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nature microbiology 1: 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrle HM, Guidry JT, Iacovetto R, Salisbury AK, Sandidge DJ, and Bowman GR (2017) Polar organizing protein PopZ is required for chromosome segregation in Agrobacterium tumefaciens. Journal of bacteriology 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kafafi el S, Mukherjee S, El-Shami M, Putaux JL, Block MA, Pignot-Paintrand I, Lerbs-Mache S, and Falconet D (2005) The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localization. The Biochemical journal 387: 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami K, Guyet A, Kawai Y, Devi J, Wu LJ, Allenby N, Daniel RA, and Errington J (2017) RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nature microbiology 2: 16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, and Osawa M (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiology and molecular biology reviews : MMBR 74: 504–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Cuilan W, Daniel JJ, Howell M, Sulaiman A, and Brown PJ (2016) Mini-Tn7 Insertion in an artificial attTn7 site enables depletion of the essential master regulator CtrA in the phytopathogen Agrobacterium tumefaciens. Applied and environmental microbiology 82: 5015–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Cuilan WM, and Brown PJB (2018) Cell wall biogenesis during elongation and division in the plant pathogen Agrobacterium tumefaciens. Current topics in microbiology and immunology [DOI] [PubMed]
- Flores SA, Howell M, Daniel JJ, Piccolo R, and Brown PJB (2018) Absence of the Min system does not cause major cell division defects in Agrobacterium tumefaciens. Frontiers in microbiology 9: 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraipont C, Alexeeva S, Wolf B, van der Ploeg R, Schloesser M, den Blaauwen T, and Nguyen-Disteche M (2011) The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology (Reading, England) 157: 251–259. [DOI] [PubMed] [Google Scholar]
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, and Xiao J (2010) In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PloS one 5: e12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, and Fukui S (1974) Effect OF D-alanine and mitomycin-c on cell morphology of Agrobacterium tumefaciens. Gen App Microbiol 20: 345–349. [Google Scholar]
- Goley ED, Dye NA, Werner JN, Gitai Z, and Shapiro L (2010) Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Molecular cell 39: 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Yeh YC, Hong SH, Fero MJ, Abeliuk E, McAdams HH, and Shapiro L (2011) Assembly of the Caulobacter cell division machine. Mol Microbiol 80: 1680–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, and Slater S (2001) Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science (New York, N.Y.) 294: 2323–2328. [DOI] [PubMed] [Google Scholar]
- Grangeon R, Zupan J, Jeon Y, and Zambryski PC (2017) Loss of PopZ At activity in Agrobacterium tumefaciens by Deletion or Depletion Leads to Multiple Growth Poles, Minicells, and Growth Defects. mBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon R, Zupan JR, Anderson-Furgeson J, and Zambryski PC (2015) PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens. Proceedings of the National Academy of Sciences of the United States of America 112: 11666–11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray AN, Egan AJ, Van’t Veer IL, Verheul J, Colavin A, Koumoutsi A, Biboy J, Altelaar AF, Damen MJ, Huang KC, Simorre JP, Breukink E, den Blaauwen T, Typas A, Gross CA, and Vollmer W (2015) Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, and Levin PA (2008) The great divide: coordinating cell cycle events during bacterial growth and division. Current opinion in microbiology 11: 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney SA, Glasfeld E, Hale C, Keeney D, He Z, and de Boer P (2001) Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. The Journal of biological chemistry 276: 11980–11987. [DOI] [PubMed] [Google Scholar]
- Holden SJ, Pengo T, Meibom KL, Fernandez Fernandez C, Collier J, and Manley S (2014) High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proceedings of the National Academy of Sciences of the United States of America 111: 4566–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M, Aliashkevich A, Salisbury AK, Cava F, Bowman GR, and Brown PJB (2017a) Absence of the polar organizing protein PopZ results in reduced and asymmetric cell division in Agrobacterium tumefaciens. Journal of bacteriology 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M, and Brown PJB (2016) Building the bacterial cell wall at the pole. Current opinion in microbiology 34: 53–59. [DOI] [PubMed] [Google Scholar]
- Howell M, Daniel J, and J.B. Brown P, (2017b) Live cell fluorescence microscopy to observe essential processes during microbial cell growth. J Vis Exp [DOI] [PMC free article] [PubMed]
- Kuru E, Velocity Hughes H, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, and VanNieuwenhze MS (2012) In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angewandte Chemie (International ed. in English) 51: 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere PJ, Szwedziak P, Mahone CR, Lowe J, and Goley ED (2018) FzlA, an essential regulator of FtsZ filament curvature, controls constriction rate during Caulobacter division. Mol Microbiol 107: 180–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latch JN, and Margolin W (1997) Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. Journal of bacteriology 179: 2373–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, and Jensen GJ (2007) The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. The EMBO journal 26: 4694–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, and Margolin W (1999) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. Journal of bacteriology 181: 7531–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W, and Long SR (1994) Rhizobium meliloti contains a novel second homolog of the cell division gene ftsZ. Journal of bacteriology 176: 2033–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, and Rudner DZ (2016) SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537: 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier EL, and Goley ED (2014) Form and function of the bacterial cytokinetic ring. Current opinion in cell biology 26: 19–27. [DOI] [PubMed] [Google Scholar]
- Meier EL, Razavi S, Inoue T, and Goley ED (2016) A novel membrane anchor for FtsZ is linked to cell wall hydrolysis in Caulobacter crescentus. Mol Microbiol 101: 265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll A, and Thanbichler M (2009) FtsN-like proteins are conserved components of the cell division machinery in proteobacteria. Mol Microbiol 72: 1037–1053. [DOI] [PubMed] [Google Scholar]
- Monahan LG, Hajduk IV, Blaber SP, Charles IG, and Harry EJ (2014) Coordinating bacterial cell division with nutrient availability: a role for glycolysis. mBio 5: e00935–00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton ER, and Fuqua C (2012a) Laboratory maintenance of Agrobacterium. Current protocols in microbiology Chapter 1: Unit3D 1. [DOI] [PMC free article] [PubMed]
- Morton ER, and Fuqua C (2012b) Unit 3D.2 genetic manipulation of Agrobacterium. Current protocols in microbiology Chapter: Unit-3D.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, and Somers WS (2000) The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. The EMBO journal 19: 3179–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller FD, Raschdorf O, Nudelman H, Messerer M, Katzmann E, Plitzko JM, Zarivach R, and Schuler D (2014) The FtsZ-like protein FtsZm of Magnetospirillum gryphiswaldense likely interacts with its generic homolog and is required for biomineralization under nitrate deprivation. Journal of bacteriology 196: 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson BJ, Wang Q, and Osteryoung KW (2010) GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. The Journal of biological chemistry 285: 20634–20643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C, Natale P, Cueto L, and Vicente M (2016) The keepers of the ring: regulators of FtsZ assembly. FEMS microbiology reviews 40: 57–67. [DOI] [PubMed] [Google Scholar]
- Pichoff S, and Lutkenhaus J (2007) Identification of a region of FtsA required for interaction with FtsZ. Mol Microbiol 64: 1129–1138. [DOI] [PubMed] [Google Scholar]
- Pini F, De Nisco NJ, Ferri L, Penterman J, Fioravanti A, Brilli M, Mengoni A, Bazzicalupo M, Viollier PH, Walker GC, and Biondi EG (2015) Cell cycle control by the master regulator CtrA in Sinorhizobium meliloti. PLOS Genetics 11: e1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pini F, Frage B, Ferri L, De Nisco NJ, Mohapatra SS, Taddei L, Fioravanti A, Dewitte F, Galardini M, Brilli M, Villeret V, Bazzicalupo M, Mengoni A, Walker GC, Becker A, and Biondi EG (2013) The DivJ, CbrA and PleC system controls DivK phosphorylation and symbiosis in Sinorhizobium meliloti. Mol Microbiol 90: 54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DM, Hempel AM, Flardh K, Buttner MJ, and Howard M (2012) Mechanistic basis of branch-site selection in filamentous bacteria. PLoS computational biology 8: e1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett VW, and Margolin W (2015) The Min system and other nucleoid-independent regulators of Z ring positioning. Frontiers in microbiology 6: 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan K, Miguel A, Desmarais SM, Meier EL, Huang KC, and Goley ED (2015) The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nature communications 6: 7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SM, and Lowe J (2012) FtsA forms actin-like protofilaments. The EMBO journal 31: 2249–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush AD, MacCready JS, Chen C, Ducat DC, and Osteryoung KW (2018) Conserved dynamics of chloroplast cytoskeletal FtsZ proteins across photosynthetic lineages. Plant physiology 176: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, and Young KD (2004) FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. Journal of bacteriology 186: 6768–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan S, Wickstead B, Gull K, and Addinall SG (2004) Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. Journal of molecular evolution 58: 19–29. [DOI] [PubMed] [Google Scholar]
- Ward JE Jr., and Lutkenhaus J (1985) Overproduction of FtsZ induces minicell formation in E. coli. Cell 42: 941–949. [DOI] [PubMed] [Google Scholar]
- Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF Jr., Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D Sr., Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, and Nester EW (2001) The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science (New York, N.Y.) 294: 2317–2323. [DOI] [PubMed] [Google Scholar]
- Wu LJ, and Errington J (2004) Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117: 915–925. [DOI] [PubMed] [Google Scholar]
- Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, and Xiao J (2017) GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science (New York, N.Y.) 355: 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan JR, Cameron TA, Anderson-Furgeson J, and Zambryski PC (2013) Dynamic FtsA and FtsZ localization and outer membrane alterations during polar growth and cell division in Agrobacterium tumefaciens. Proceedings of the National Academy of Sciences of the United States of America 110: 9060–9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


