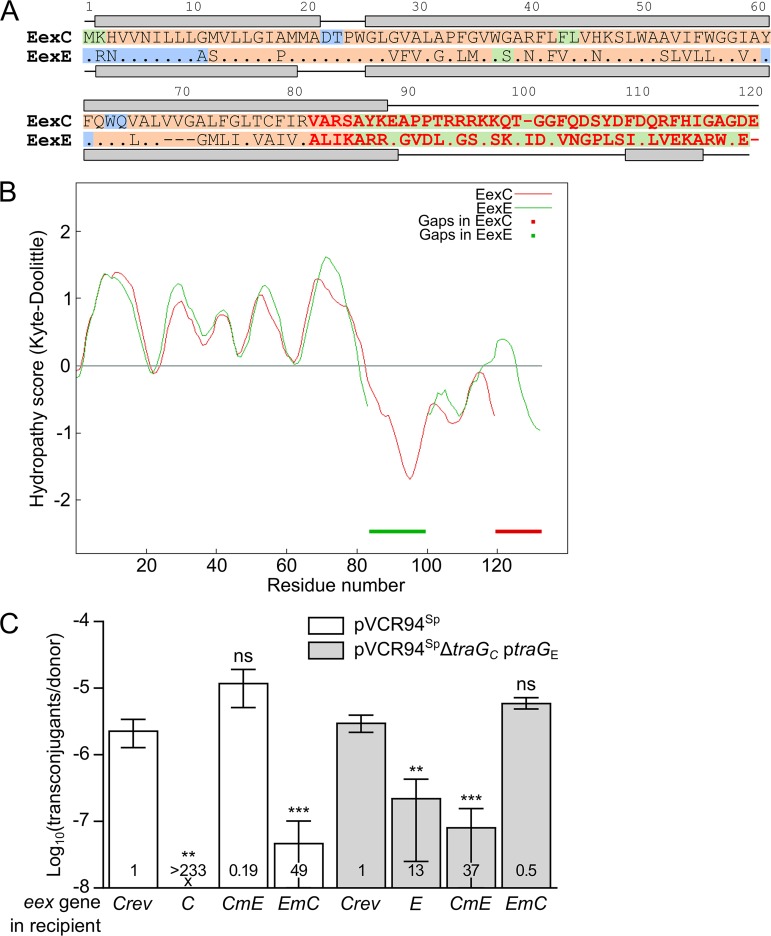
FIG 4.
Localization of the specificity domain of Eex proteins. (A) MUSCLE alignment of EexC and EexE. EexC was used as the reference, and identical amino acids in EexE are represented by a dot. Predicted secondary structures are shown above and below the sequence of each protein, with lines and gray boxes representing coils and helices, respectively. Background colors indicate amino acid residues predicted to be cytoplasmic (green) or periplasmic (blue) or present within a transmembrane helix (orange). Variable C termini of EexC and EexE shown in red were swapped to construct the chimeric proteins CmE and EmC. (B) Aligned Kyte-Doolittle hydropathy plots of EexC and EexE. Thick lines indicate alignment gaps in EexC and EexE. (C) Specificity of Eex chimeras toward TraGC and TraGE. Mating assays were performed using as the donor strains GG56 bearing either pVCR94Sp or its ΔtraGC mutant with ptraGE. All recipient strains were derivatives of CAG18439 expressing eexC or eexE or either of the two chimeras eexCmE and eexEmC. CAG18439 bearing peexCrev was used as the nonexclusion control. Each bar represents the mean of results from three independent experiments, with error bars indicating the standard deviations. Exclusion indexes (EI) are indicated at the bottom of each bar. One-way ANOVAs with Dunnett's multiple-comparison test were carried out on the logarithm of the values to compare each bar to the nonexclusion control for each donor. ***, P < 0.001; **, P < 0.01; ns, not significant.