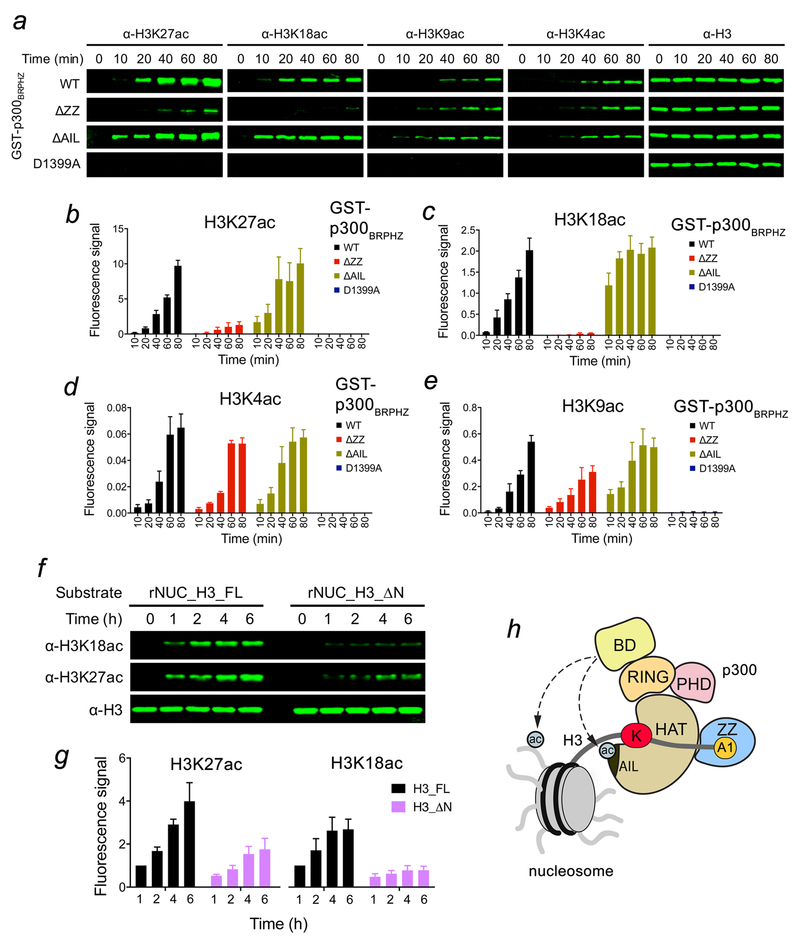
Figure 4. p300 ZZ-H3 interaction promotes acetylation of H3K27 and H3K18.
(a) In vitro HAT assays using p300BRPHZ, wild type and counterparts harboring either ZZ deletion, AIL deletion or loss-of-catalytic activity HAT mutation (D1399A), and the reconstituted nucleosome. (b-e) Quantification of the HAT activity based on the fluorescence signal from three biological replicates. A common standard sample is used for normalization in each replicate. (f) In vitro HAT assays using WT p300BRPHZ and the reconstituted nucleosome carrying either intact H3.1 (rNUC_H3_FL) or N-terminally truncated H3.1 (rNUC_H3_ΔN, Ala1 and Arg2 of H3.1 are deleted). (g) Quantification of the HAT activity on H3K18 and H3K27 based on the fluorescence signal in (f) from three biological replicates. The 1 hr sample of rNUC_H3_FL is used for normalization. (h) A model for the regulation of the p300 HAT activity by the BD and ZZ. Potential intermolecular interactions of BD with acetylated histones and acetylated AIL are indicated by dashed lines. Error bars in (b-e, g) represent s.e.m. from 3 independent HAT assays using different batches of enzymes (n = 3 biological replicates). Uncropped blot images (a, f) are shown in Supplementary Data Set 1.