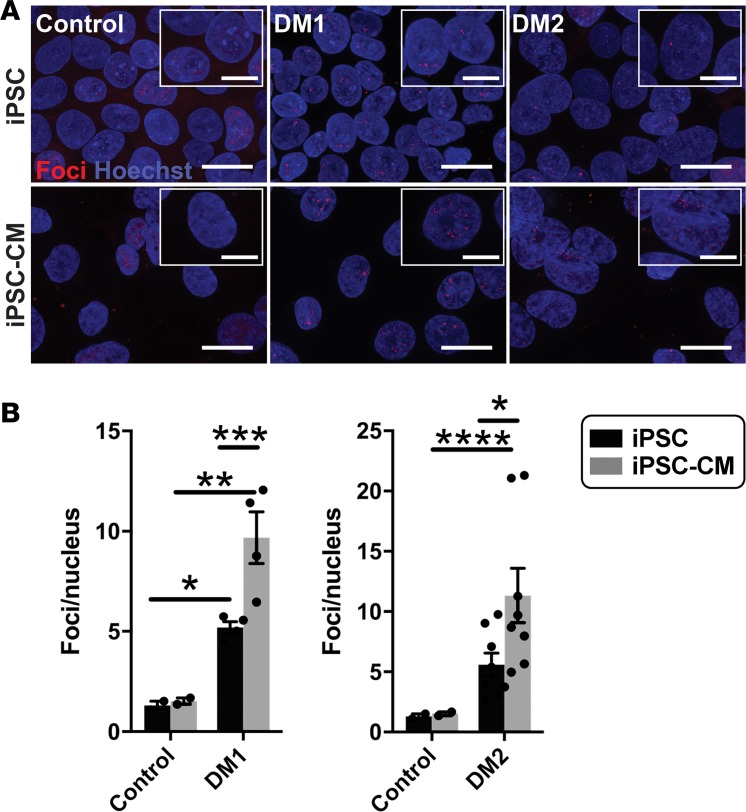
Figure 6. Fluorescence in situ hybridization (FISH) detected an increase in RNA foci after cardiomyocyte differentiation.
FISH was used to detect RNA-encoded nucleotide repeat expansions using probes specific to the repeat expansions in DM1 or DM2. RNA probes for FISH included either (CAG)10 to detect DM1 or (CAGG)5 to detect the DM2 repeat expansion. Probes were labeled with Cy3 (red), FISH was conducted, and the number of foci was quantified and compared in undifferentiated iPSCs and iPSCs that had been differentiated to cardiomyocytes (iPSC-CM). (A) Example images of RNA foci (red) visualized using Cy3-labeled probes specific for the myotonic disease subtype. Nuclei were labeled with Hoechst (blue). Magnified images are shown in the insets. Scale bar: 20 μm; 10 μm (inset). (B) DM1 iPSCs and iPSC-CMs had an increased number of RNA foci compared with healthy control cells (*P = 0.04, **P = 0.0008, respectively). DM2 iPSC-CMs had an increased number of RNA foci compared with healthy control cells (****P = 0.03), while iPSC cells trended toward significance when compared with control cells (P < 0.07). For both DM1 and DM2, differentiation of iPSCs into iPSC-CMs resulted in an increase number of RNA repeat foci in iPSC-CMs (***P = 0.008, *P = 0.04, respectively). The number of RNA foci did not change with differentiation in control cells (2-way ANOVA).