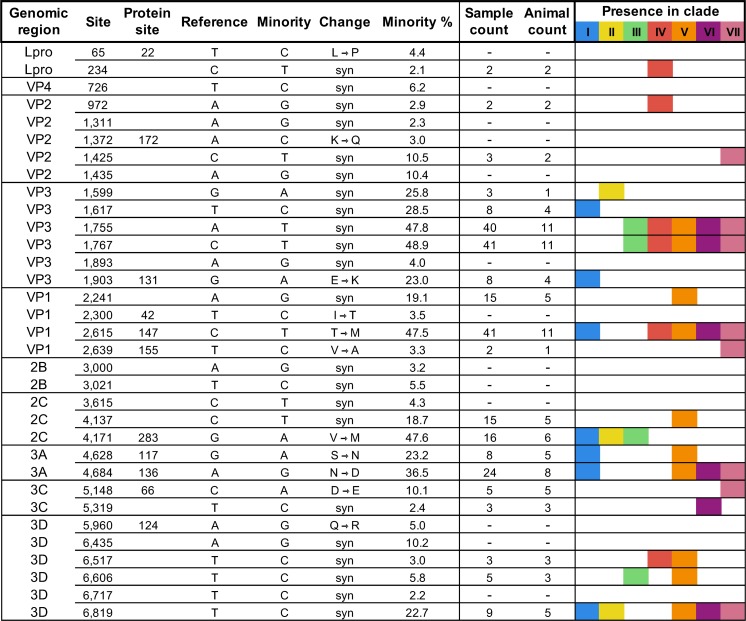
Fig 1. Subconsensus variants present above 2% frequency in inoculum ultra-deep sequencing.
Reference is the consensus sequence of the inoculum and minority represents the polymorphic base at each site. Nucleotide variation was not detected in inoculum 2A and 3B1-3 coding regions. Sample count indicates the number of sample consensus sequences (of 52 total) that encoded each nucleotide found at the minority (2–49%) level in the inoculum deep sequence. Animal count indicates the number of individual hosts that provided sequences with these variants of the 13 total. If no value is indicated (-), no sample sequence encoded the inoculum’s minority nucleotide at the consensus level. Roman numerals and colors indicate the presence of each specific site change in at least one clade member as described in Results: Phylogenetic Associations.