Figure 1.
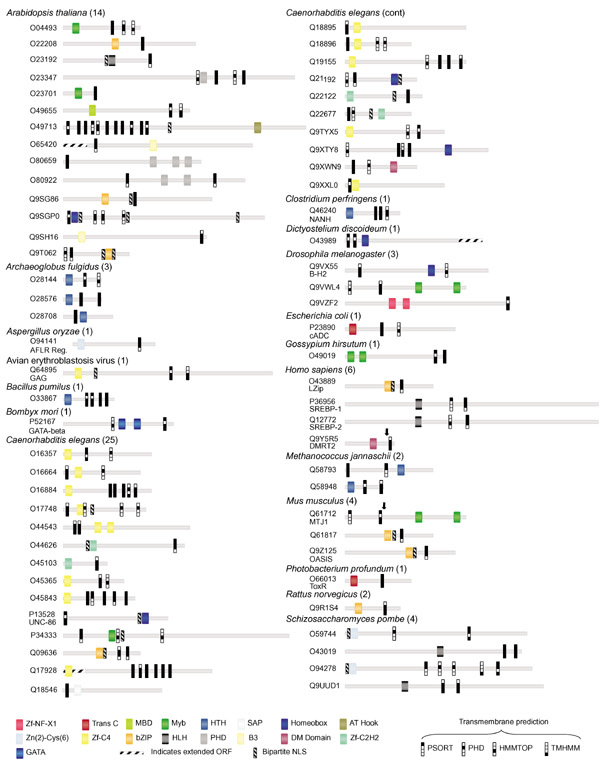
The domain structure of predicted TMTFs is shown. Pfam-predicted DNA-binding domains, transmembrane segments and bipartite nuclear localization signals are shown for linear protein models and identified by SWISS-PROT/TrEMBL accession number. The total number of proteins predicted for each species is given. Colored icons represent various DNA-binding domains. Predicted transmembrane segments for each program are represented by a filled box. Protein lengths are drawn approximately to scale; positions of domains are approximate. Arrows in MTJ1 and DMRT2 indicate sites for truncated protein localization experiments shown in Figure 2. The scale of proteins O80659 and Q9SGP0 is reduced by half. Orthologs of predicted TMTFs not shown are: Luman (Q9UE77 Homo sapiens), SREBP-1 (Q60416 Cricetulus griseus, Q9WTN3 Mus musculus, P56720 Rattus norvegicus, Q9XX00 Caenorhabditis elegans), SREBP-2 (Q9UH04 H. sapiens, Q60429 C. griseus), and AFLR Reg (P43651 Aspergillus parasiticus). Open reading frames (ORFs) for O65420, O43989, Q17928 were extended using additional nucleotide sequence available in the NCBI database (indicated by stippled rectangles).
