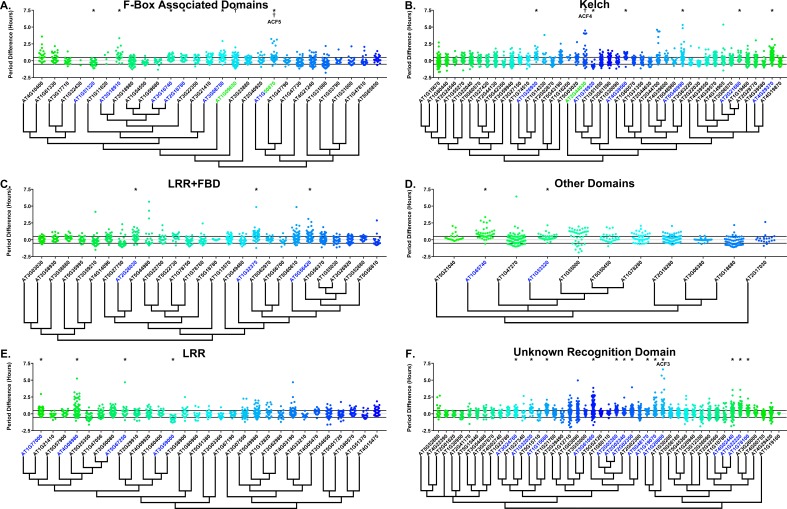
Figure 6. Period distributions of F-box decoy plants.
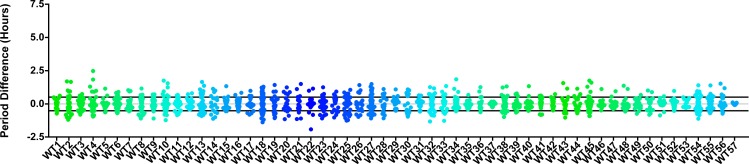
Values presented are the difference between the period of the individual decoy plant and the average period of the CCA1p::Luciferase control in the accompanying experiment. The grey line is at the average control value and the black lines are at ±the standard deviation of the control plants. Genes are separated by protein recognition domain and ordered by closest protein homology using Phylogeny.Fr, (Dereeper et al., 2008), and a tree showing that homology is displayed beneath the graph. F-Box Associated Domains = FBA1, FBA3, and FBD only. Other Domains = TUB, JmjC, LysM, WD40, zf_MYND, and DUF295. * and blue gene names = The entire population differs from wildtype with a Bonferroni-corrected p<2.55×10−4; † and green gene names = A subset of the population differs from wildtype with a Bonferroni-corrected p<2.55×10−4. Only data from those experiments where the control CCA1p::Luciferase plants display a standard deviation less than 0.75 were included in our analyses.