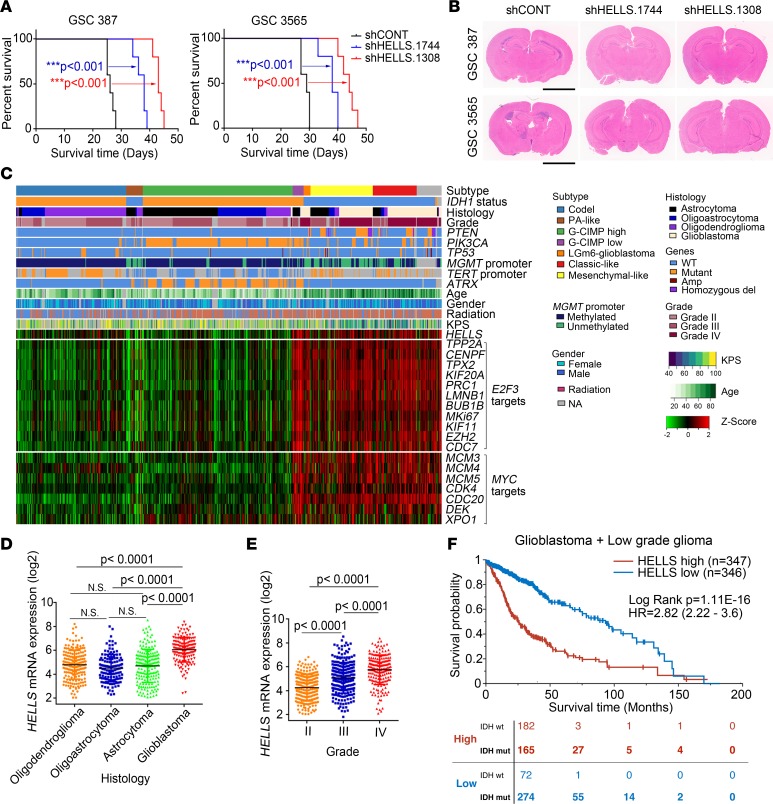
Figure 9. HELLS is required for in vivo glioblastoma growth and informs prognosis.
(A) Kaplan-Meier survival curves of immunocompromised mice bearing intracranial GSC 387 and 3565 transduced with either 1 of 2 shRNAs targeting HELLS or nontargeting control. ***P < 0.001, by Mantel-Cox log-rank test. n = 5. (B) Representative images of H&E-stained brain sections 20 days after transplanting GSC 387 and 3565 transduced with either 1 of 2 shRNAs targeting HELLS or nontargeting control. Scale bars: 3 mm. (C) RNA-seq, whole exome, and clinical phenotype data were aggregated from TCGA glioblastoma (GBM) and low-grade glioma (LGG) data sets to visualize the expression patterns of HELLS, E2F3 targets, and MYC targets across glioma. “Codel,” codeletion of chromosomes 1p and 19q; PA-like, pilocytic astrocytoma–like; CIMP, glioma-CpG island methylator phenotype; LGm6-GBM, a subgroup of glioma enriched for histologic low-grade gliomas that also contains a subset of tumors with GBM-defining histologic criteria; KPS, Karnofsky performance status. (D) HELLS expression in patients with oligodendroglioma, oligoastrocytoma, astrocytoma, and glioblastoma. Data are presented as mean ± SD. P values were determined by 1-way ANOVA with Tukey’s multiple comparisons test. (E) HELLS expression across glioma tumor grades. Data are presented as mean ± SD. P values were determined by 1-way ANOVA with Tukey’s multiple comparisons test. (F) Survival curves of glioma patients with higher and lower HELLS expression. Significance was determined by Mantel-Cox log-rank testing. The distribution of IDH WT and mutant patients in both the HELLShi (red) and the HELLSlo (blue) group at the indicated time points is listed at bottom.