Figure 2: m1G37 deficiency in E. coli and Salmonella affects cell viability.
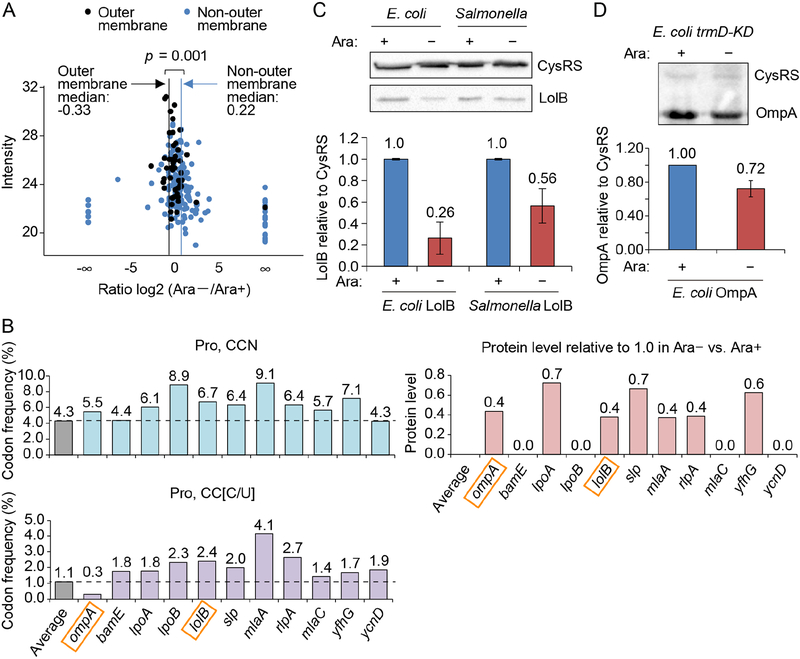
A) Quantitative mass spectrometry analysis of membrane proteins in E. coli trmD-KD cells isolated from Ara− and Ara+ conditions. The label-free quantification intensity is compared to the signal of log2 (fold-change) (Ara−/Ara+). OM proteins are plotted in black with a vertical line indicating the median of −0.33 (equivalent to a decrease of 21%), while non-OM proteins are plotted in blue with a vertical line showing the median of 0.22 (equivalent to an increase of 16%). p < 0.001 by a Kolmogorov-Smirnov analysis.
B) Frequency of Pro codons CCN (top) and CC[C/U] (bottom) in genes whose OM proteins are reduced in Ara− vs. Ara+ in (A). Each frequency is compared to the average frequency of respective Pro codons in E. coli protein-coding genes.
C,D) m1G37 deficiency decreased LolB levels (C) to 26% in E. coli and to 56% in Salmonella, and decreased OmpA levels in E. coli to 72% (D) in Western blots (top). Overnight cultures of trmD-KD cells were diluted 1:100 into fresh LB with or without 0.2% Ara and grown for 4 h at 37 °C. Cells were inoculat ed into fresh LB in Ara+/− conditions for another 3 h. Data and error bars represent mean ± SD, n = 4.
See also Figure S3.