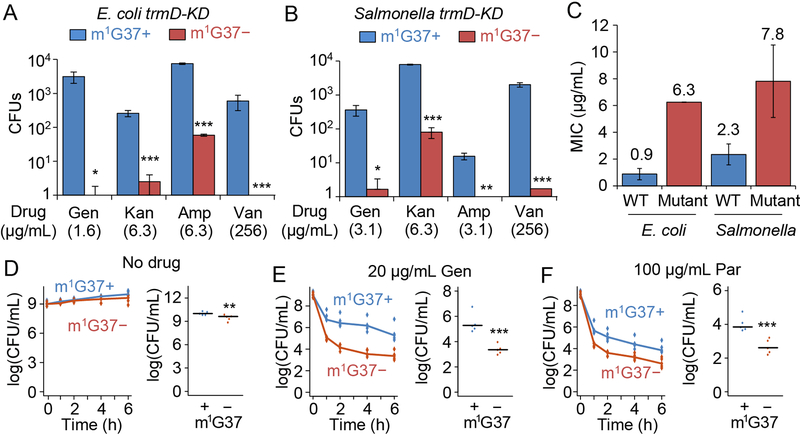
Figure 5: m1G37 deficiency decreases resistance and persistence to antibiotic treatment.
A-C) Resistance arises less frequently in m1G37− (red) E. coli (A) and Salmonella (B) trmDKD cells than in m1G37+ (blue) cells. An overnight culture of cells at 105 CFUs was plated onto an LB agar plate containing the indicated concentration of gentamicin (Gen), kanamycin (Kan), ampicillin (Amp), or vancomycin (Van). Each concentration was near 1X MIC for m1G37+ cells. Resistant colonies were counted after incubation at 37 °C for 3 days. Mutants were verified to have an increase in MIC to the respective antibiotic (C). Data and error bars are mean ± SD, n = 3. Welch’s t-test: *p < 0.1, **p < 0.05, ***p < 0.01.
D-F) Persistence of Salmonella trmD-KD cells, showing CFUs/mL over time (left) and the average CFUs/mL at 6 h post-treatment in m1G37+ and m1G37− conditions (right). Untreated Salmonella trmD-KD cells had similar CFUs/mL in the two conditions (D), while persistence arose more frequently in m1G37+ than m1G37− cells treated with 20 μg/mL Gen (3.7X and 8.5X MIC for m1G37+ and m1G37− conditions) (E) and with 100 μg/mL paromomycin (Par; 2.7X and 10.7X MIC for m1G37+ and m1G37− conditions) (F). An overnight culture in LB with 0.2% Ara was diluted 1:100 into fresh LB with or without 0.2% Ara and incubated at 37 °C for 3 h. Cells were treated with water (no drug), Gen or Par for 0, 1, 2, 4, and 6 h, collected, washed, and plated on LB with Ara. Horizontal lines on the right represent the median, n = 5. Mann-Whitney U test: **p < 0.05, ***p < 0.01.