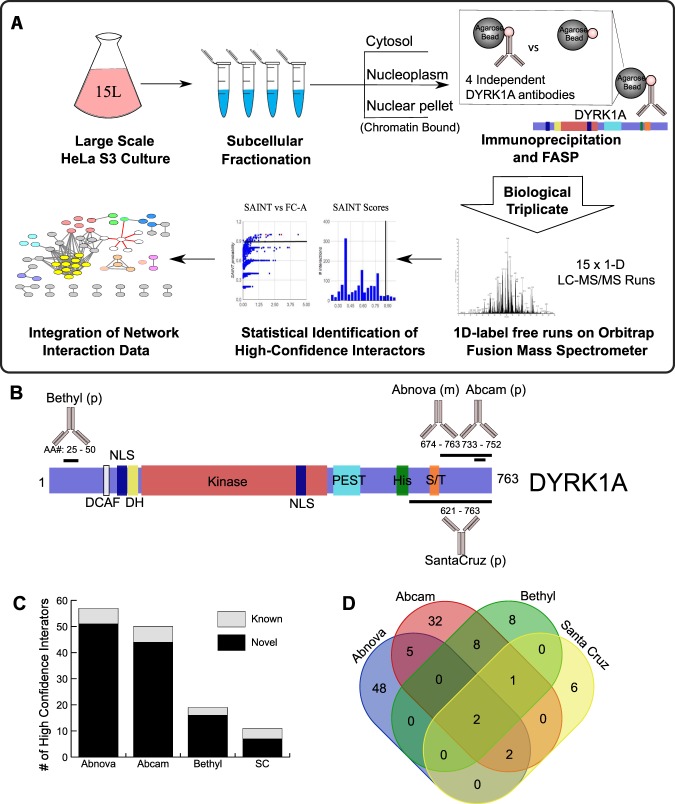
Figure 1.
Workflow and strategy for generation of endogenous DYRK1A interactome. (A) Schematic overview of fractionation and IPMS strategy. HeLa S3 cells were grown in 6 L flasks before harvesting at 1 million cells/ml. Large scale fractionation of cell pellets generated protein lysates representing a cytosolic, nuclear and chromatin bound fractions. Subsequent immunoprecipitations were done in triplicate prior to sample preparation for mass spectrometry identification. (B) Domain arrangement of DYRK1A and regions containing antibody epitope as reported by manufacturers. NLS, nuclear localization signal; DH, DYRK homology box; PEST, proline glutamic acid serine and threonine rich sequence; His, polyhistidine stretch; S/T serine and threonine rich region. Epitopes recognized by each antibody used for IP experiments fall within the designated amino acid region and represented by black bar. (p) rabbit polyclonal; (m) mouse monoclonal. (C) Number of novel and literature reported HCI’s by antibody. (Abnova n = 3; Abcam n = 3; Bethyl n = 3; SC: Santa Cruz n = 3) FC-A > 3; SAINT > 0.7. (D) Overlap between HCI’s of each set of immunoprecipitations by antibody.