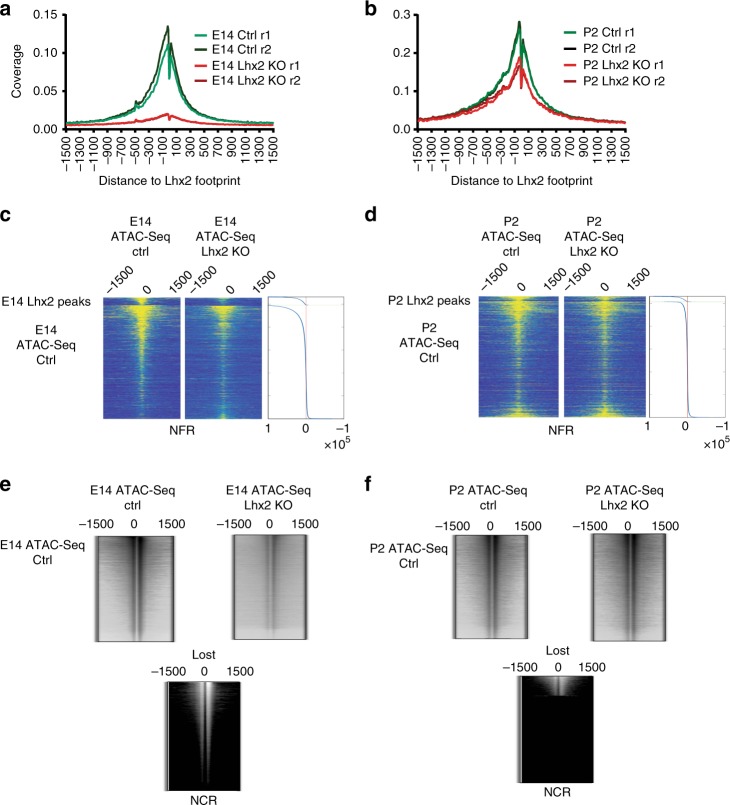
Fig. 4.
LHX2 affects local and global chromatin accessibility. a, b ATAC-Seq read distribution was plotted relative to the center of LHX2 footprints from controls and age-matched Lhx2 cKO. c, d Read distribution profiles across nucleosome free regions (nfr) are derived from ATAC-Seq control and Lhx2 cKO, plotted around age-matched LHX2 ChIP-Seq peaks centers and across all the identified age-matched open chromatin regions in a 3 kb window. The densitometric difference in read coverage between control and Lhx2 cKO is reported on the right. e, f Read distribution profiles flanking nucleosomes centered regions (ncr) are derived from ATAC-Seq control and Lhx2 cKO and plotted across all the identified age-matched open chromatin regions in a 3 kb window