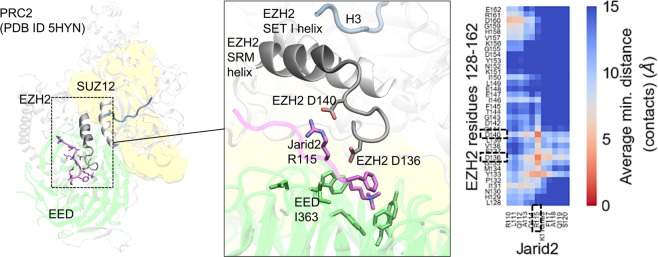
Figure 1.
Structure and dynamics of PRC2 activation. Crystal structures of PRC210 (left and zoomed in Jarid2 peptide binding site) and supporting MD simulations reveal the mechanism of PRC2 activation: upon EED (green) binding Jarid2 R115 (magenta) forms a salt bridge with D140 (and to a lesser extend with D136) on EZH2 loop stabilizing the conformation of EZH2 SRM helix (gray) which then in turn stabilizes EZH2 SET-I helix (white) of the catalytic binding site for substrate H3K27 (blue). The heatmap plot represents the average minimum residue-residue distances (contacts) between EZH2 and Jarid2 peptide highlighting the importance of the salt bridge, as determined from cumulative 2.5 µs MD sampling (SI Fig. S2). For clearer representation the labels on EED aromatic cage residues (F97, Y148, W364, Y365) and D362 as well as Jarid2’s K116me3 and F117 are hidden.