Figure 4.
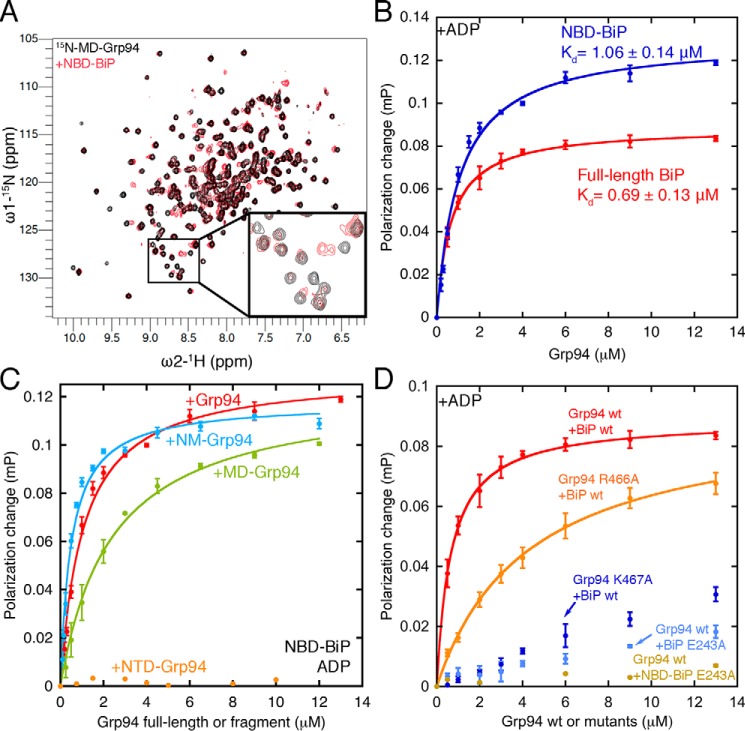
A, direct binding between the BiP NBD and Grp94 MD shown by chemical shift perturbations ([15N]Grp94 MD alone, black, 150 μm, 32 scans; [15N]Grp94 MD with BiP NBD, red, 200 μm, 32 scans). B, the BiP NBD and full-length BiP have comparable affinities for Grp94 under ADP conditions. C, the Grp94 MD is sufficient for binding the BiP NBD, but full affinity requires both the MD and NTD. D, mutants that disrupt binding between the bacterial Hsp70 and Hsp90 system also disrupt binding between BiP and Grp94. Error bars are the S.E. of the mean for at least three measurements. Buffer conditions for binding experiments are the same as in Fig. 3. mP, millipolarization units.