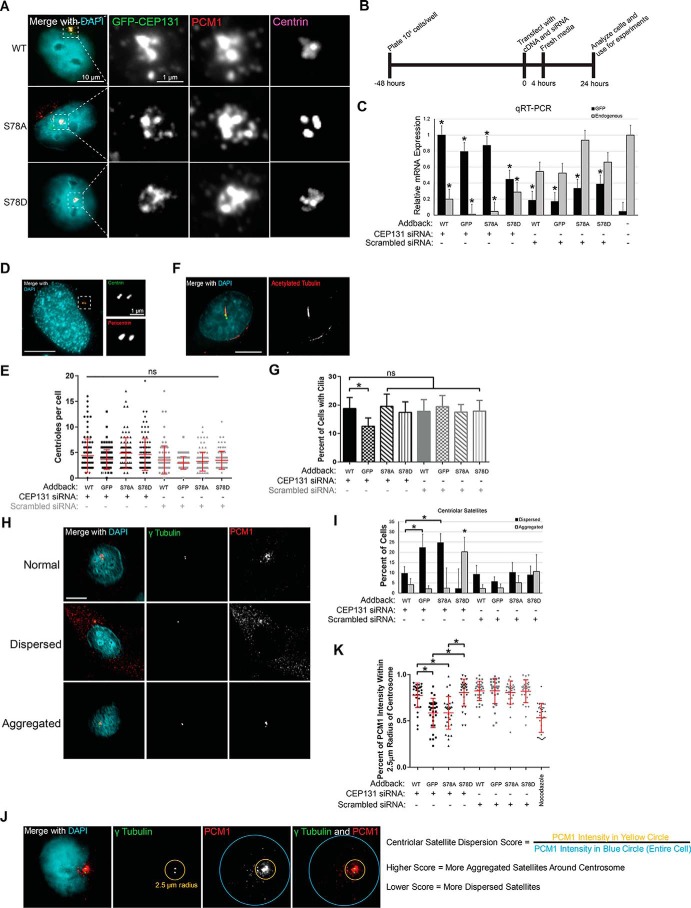
Figure 3.
PLK4 phosphorylation of CEP131 maintains centriolar satellite integrity. A, localization of WT CEP131 and S78A and S78D mutants in WT HeLa cells. The dotted white box is enlarged for the adjacent gray scale images showing GFP, PCM1, and centrin at the centrosome. Scale bar sizes are indicated on the images. B, knockdown addback experiment timeline. C, quantitative RT-PCR experiment demonstrating the efficiency of knockdown and addback of CEP131. Asterisks demonstrate significant enrichment of GFP-tagged CEP131 versus endogenous GFP (black bars) or significant depletion of endogenous CEP131 versus untreated cells (gray bars). D and E, images and quantification of centriole duplication. Each dot represents one cell, and bars represent means ± S.D. (error bars) of at least 100 cells/condition. Centrioles were identified by counting centrin foci. For example, the image in D shows two centrosomes with four total centrioles. F and G, images and quantification of ciliogenesis. Cilia were marked by staining for acetylated tubulin. At least 300 total cells were analyzed per condition. H and I, images and quantification of centriolar satellites. At least 300 total cells were analyzed per condition. J, schema of a more quantitative approach to assess centriolar satellites. The yellow circle measures the PCM1 intensity within a 2.5-μm radius of the center of the centrosome, whereas the blue circle measures the PCM1 intensity in the entire cell. K, quantification of centriolar satellites using the schema described in J. Nocodazole is used as a positive control for centriolar satellite dispersion. All experiments in this figure utilize HeLa cells. Bars, means ± S.E. (error bars) from three independent experiments. Scale bars, 10 μm. *, p < 0.05. NS, not significant.