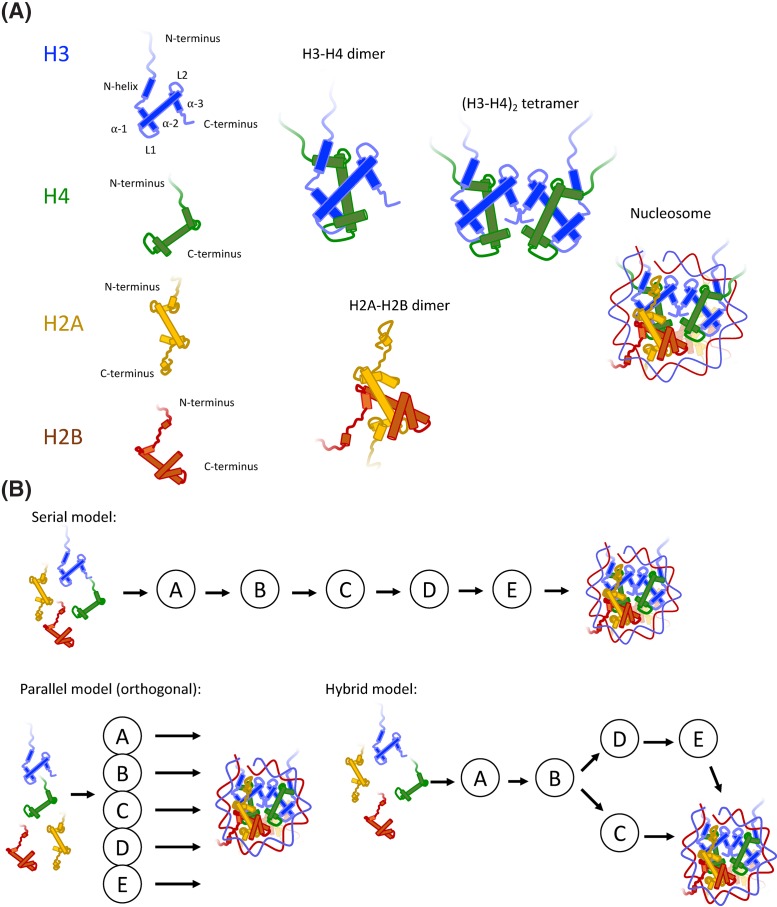
Figure 1. Components of the nucleosome and considerations for multichaperone networks.
(A) Nucleosomes comprise of four core histones – H2A, H2B, H3 and H4. The central histone fold comprises three helices, α1, α2 and α3, which can be flanked by extension on the N and C termini. N-terminal extensions are in the form of basic tails, and in the case of H3, an additional α-helix (αN-helix). H3 and H4 fold to form an H3–H4 dimer, two of which can form an (H3–H4)2-heterotetramer. The tetramer adopts the central location in the nucleosome, known as a ‘tetrasome’ when bound to DNA on its own, and is capped by two H2A–H2B dimers to form the nucleosome core particle, which wraps 145–147 base pairs of DNA in almost two superhelical turns. (B) Histones associate with a large repertoire of chaperoning complexes, represented as A, B, C, D and E. Assuming mutual exclusivity in binding, histones may transition through multiple chaperoning complexes before their incorporation in chromatin (a serial pathway), or each chaperoning complex may represent an orthologous route to deposition (a parallel pathway), or indeed, a mixture of the two (a hybrid model). In reality, a hybrid model which bifurcates depending of histone isotype best explains the H3–H4 deposition pathway (see Figure 2).