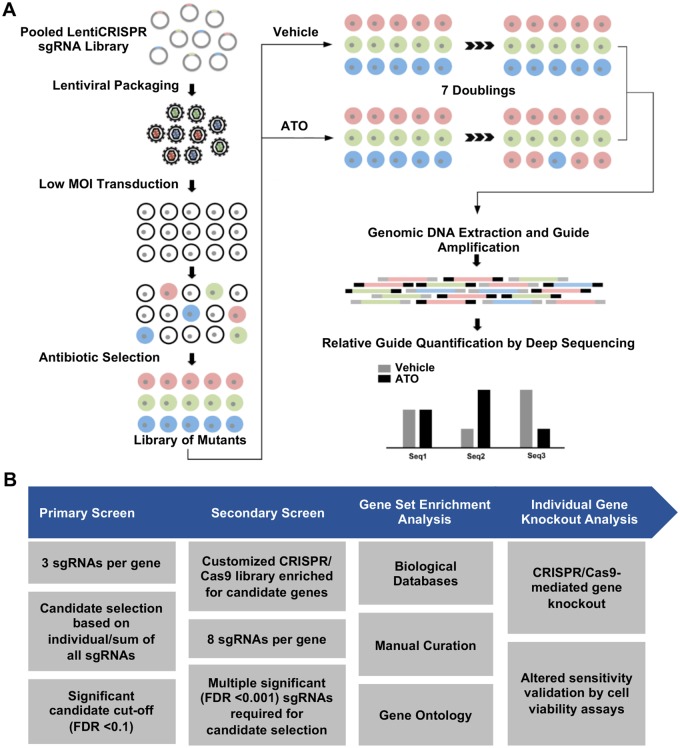
Figure 1.
Overview of the performed CRISPR-based knockout screening approach. A, The Cas9-sgRNA library was packaged into lentiviral particles and transduced into K562 cells. The obtained cellular library was screened to identify mutants with growth advantage or disadvantage in the presence of ATO. Guide sequences, used as barcodes labeling the different mutants, were PCR-amplified and quantified by deep sequencing. B, Study workflow showing screening strategies, candidate gene selection criteria, and validation approaches.