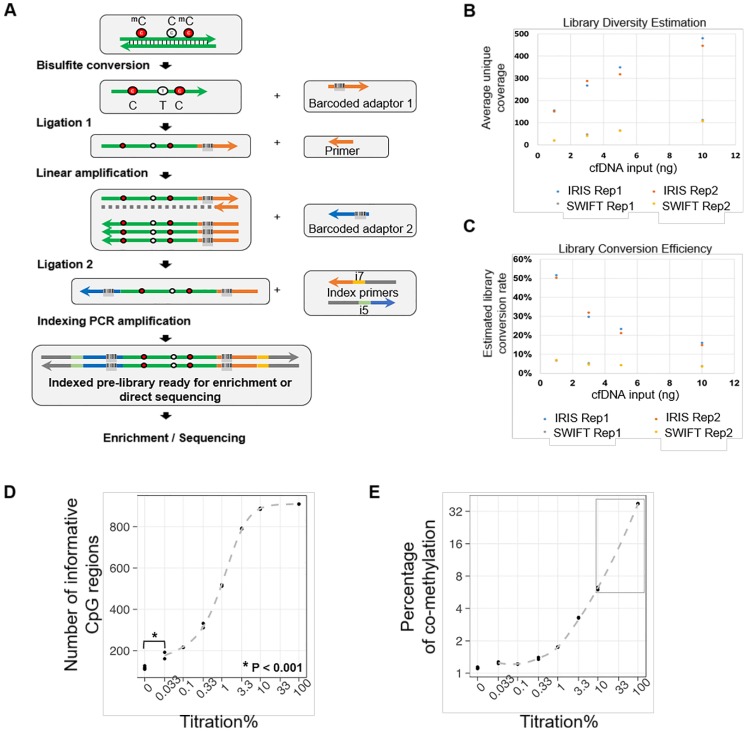
Figure 1.
The AnchorIRISTM assay and performance assessment. (A) Workflow of the ultra-sensitive AnchorIRISTM library preparation method. (B-C) A bake-off experiment comparing assay performance between the AnchorIRISTM assay and the SWIFT® accel-NGS Methyl-seqTM assay. The IRIS assay presents superior molecule conversion efficiency (C) with much higher average unique coverage for each input amount tested (B). (D and E) The sensitivity of the AnchorIRISTM assay was evaluated by diluting tumor gDNA into WBC gDNA, showing that significantly more informative co-methylated CpG regions above WBC background can be detected at dilutions ≥ 0.033% by Z-test (D). Dilutions higher than 10% (gray box) preserve a linear response of average co-methylation signal to the tumor fractions of input DNA (E).