Figure 1.
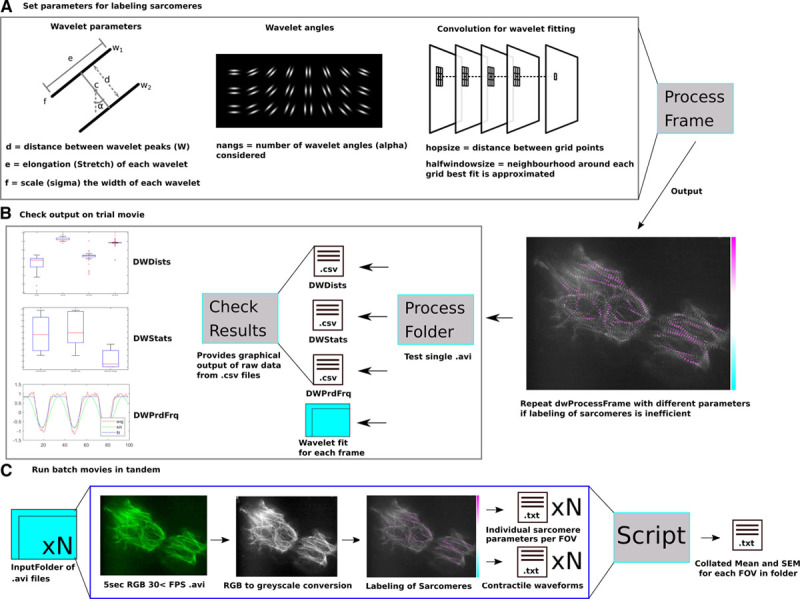
Using SarcTrack to identify fluorescent sarcomeres. A, Initial parameters are defined on a single frame of an acquired image stack to test fitting of Z-discs by SarcTrack wavelets. Wavelet parameters include the wavelet dimensions (d, e, f), the df for fitting the wavelet in certain orientations (α, the angles around c), and the definition of the grid points when performing the convolution for fitting the wavelets to fluorescent Z-disc pairs (hopsize and halfwindowsize). Once parameters are defined (see Online Methods), dwProcessFrame can be run to visualize the goodness of wavelet fitting to a specific frame. This process is repeated to find the best parameters for labeling Z-discs. In this example, we show a highly disordered cell with significant out of focus fluorescence to demonstrate the ability of these parameters to be adapted to robustly define true Z-disc pairs. B, Once parameters have been defined and tested on a single frame a stack can be analyzed using dwProcessFolder. This process allows the user to deposit one stack into a folder for analysis and this provides 4 outputs. A folder containing each individual frame fit with marked wavelets to check fidelity of wavelet fitting, DWPrdFrq providing the period and frequency of beating defined by sinusoidal fitting of the contractile functions, DWDists providing the distance (d, in pixels) of each wavelet pair for each frame of the stack. DWStats provides the precalibrated raw values derived from the sawtooth fitting of contractile cycles of each sarcomere tracked including contraction time (frames), relaxation time (frames), the minimum distance between wavelets, and the maximum distance between wavelets (pixels). dwCheckResults will output graphs of these parameters and show the fitting of the sinusoid and sawtooth for the combined sarcomere contractility of the entire field of view (FOV). C, Once parameters have been set and quality control of data has been approved using dwCheckResults one can run a folder of multiple movies. A computing cluster can be used to run many movies in parallel, and by using 4 cores, users can analyze 5-second movies every 40 min with parallel batches of up to 100 movies at a time. Once movies are batch analyzed, a script is applied to convert the outputs from DWStats to microns and seconds from pixels and frames. A mean and SEM is obtained from each DWStats file and data is output into a single .txt file. This script also adds the number of sarcomeres tracked in each movie as a separate column to all other parametric means and errors. This workflow ensures that the user can efficiently troubleshoot wavelet fitting and batch analyze hundreds of movies and tens of thousands of sarcomeres in a few hours.
