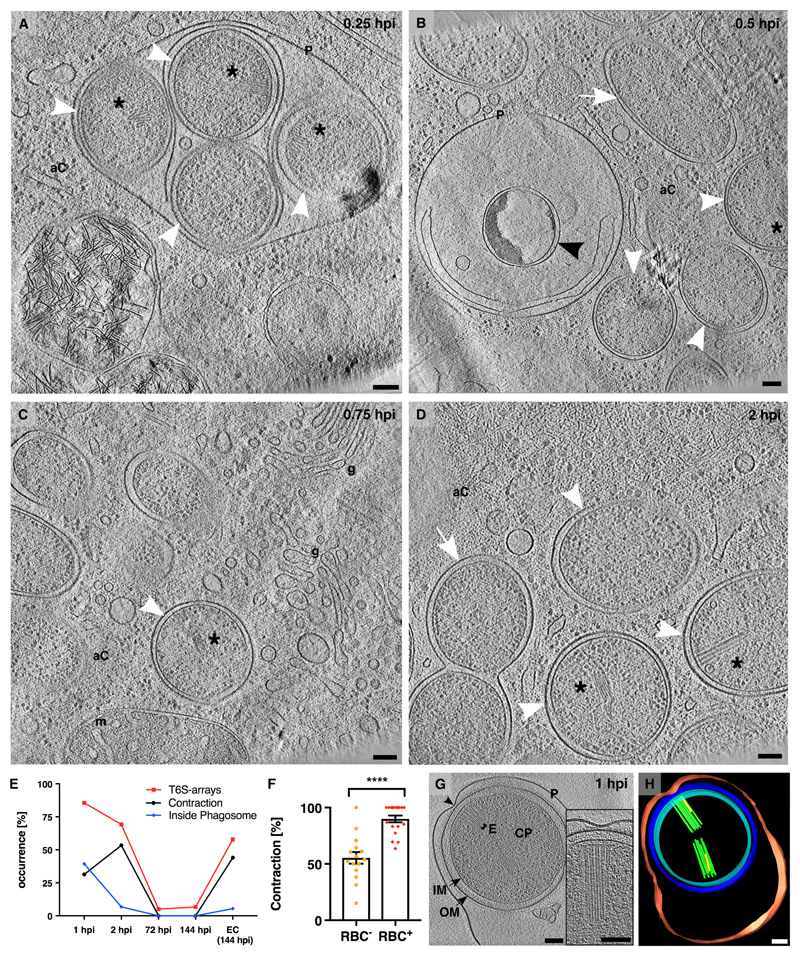
Figure 3. T6S arrays are required during early infection stages and mediate interactions with host membranes.
(A-D) Bacteria inside their host were imaged by cryo-focused ion beam milling and ECT (fig. S5). At 0.25 hpi, most coccoid amoebophili (white arrowheads) were found inside phagosomes (“P”). At later time points (0.5-2 hpi), amoebophili had escaped into the amoeba cytosol (“aC”). Amoebophili differentiated into rods (white arrows) and divided. A small fraction did not escape, showing signs of degradation (black arrowhead). Shown are 15-nm slices through cryotomograms. Asterisk, T6S array; “g”, golgi apparatus; “m”, mitochondrion. Bars: 100 nm. (E) Abundance of T6S arrays was determined by ECT of amoebophili purified from synchronized cultures, and found to be highest in extracellular “EC (144 hpi)” and early intracellular infection stages (1 hpi, 2 hpi). The increase of the contraction rate between 1 and 2 hpi correlated with the escape from the phagosome. Shown are the percentages of cells with T6S arrays (red), percentages of T6S structures that were contracted (black), and percentages of cells found inside phagosomes (blue). T6S arrays, number of quantified amoebophili: n1hpi=25, n2hpi=13, n72hpi=20, n144hpi=15, nEC(144 hpi)=19; Contraction, number of quantified T6S structures: n1hpi=168, n2hpi=88, n72hpi=4, n144hpi=4, nEC(144 hpi)=59; Inside Phagosome, number of quantified amoebophili: n1hpi=121, n2hpi=118, n72hpi=218, n144hpi=337, nEC(144 hpi)=55). (F) Amoebophili showed hemolytic activity (fig. S7). Extracellular amoebophili that interacted with RBCs showed an increased T6S contraction rate (**** p < 0.0001; nRBC–=506; nRBC+=480). (G/H) Amoebophili residing in phagosomes revealed contact sites (black arrowhead) between the Amoebophilus outer membrane and the phagosome. Any such contact site correlated with a T6S array (n=14). Shown are a 15-nm tomographic slice (G) and the corresponding model (H). “P”/red, phagosome; “OM”/blue, outer membrane; “IM”/cyan, inner membrane; “CP”, cytoplasm; “E”/green, extended T6SS; yellow, contracted T6SS; Bars: 100 nm.