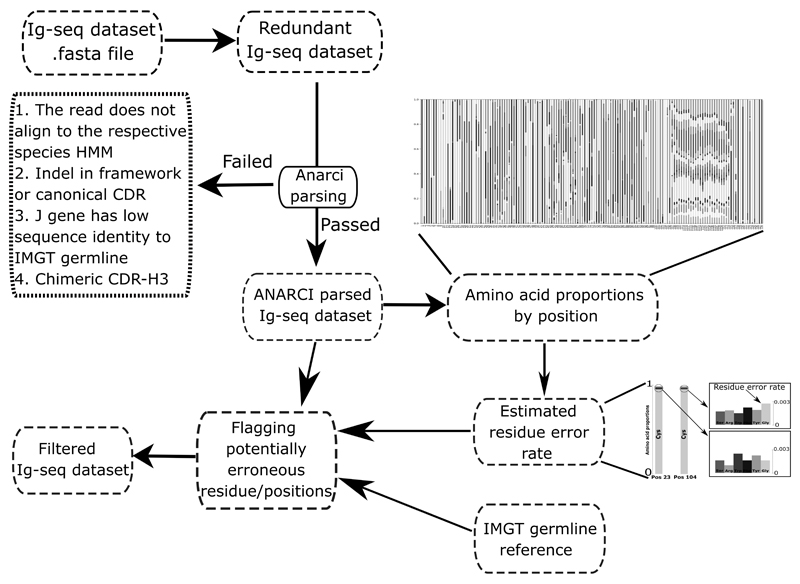
Figure 2. Workflow of ABOSS.
ABOSS input is antibody amino acid sequences in the FASTA format. Every sequence from the input file is IMGT-numbered with ANARCI (ANARCI parsing). The amino acid distribution by IMGT position is calculated for successfully ANARCI parsed sequences. The residue error rate is estimated based on the amino acid distributions at positions 23 and 104 (see Figure 1 for more details). The estimated residue error rate together with the ANARCI numbered IMGT germline genes are used to flag potentially erroneous residue/positions in individual Ig-seq sequences. Filtered Ig-seq dataset refers to a collection of sequences that pass ABOSS analysis with zero flagged residues/positions.