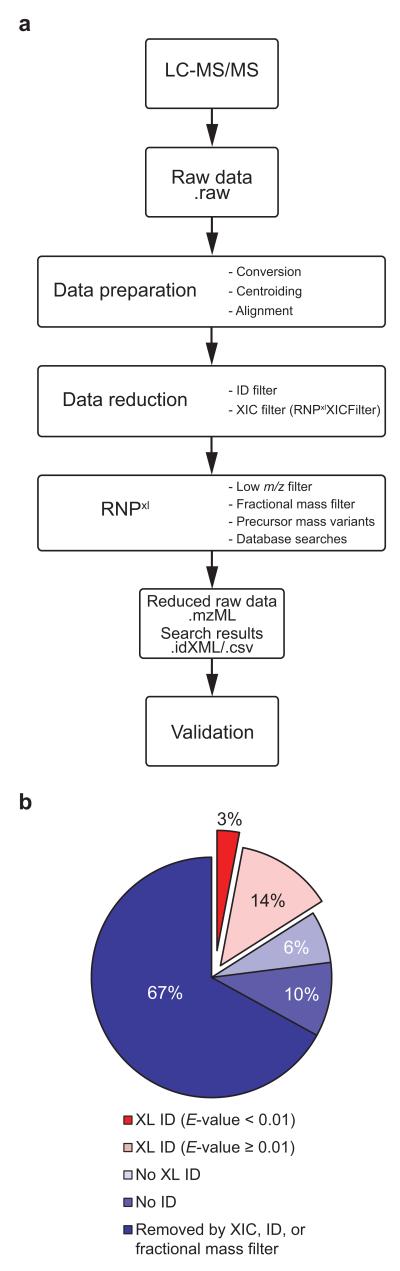
Figure 2.
Data analysis workflow and achieved data reduction in representative datasets. (a) Outline of data analysis pipeline. Raw data from LC-ESI-MS/MS experiments is submitted to the data analysis pipeline. In preparation for subsequent steps, raw data is converted into an open mass spectrometry format (.mzML) and centroided; retention time alignment between the UV sample and the nonirradiated control is performed. Next, the overall amount of data is reduced by removing MS/MS spectra of confidently identified noncross-linked peptides (ID filter) and species appearing in both UV-irradiated sample and control with comparable intensities (XIC filter with RNPxlXIC). Lastly, the RNPxl tool removes MS/MS spectra with small precursors masses (low m/z filter) and residual short oligonucleotides (fractional mass filter). For the key steps in data analysis, RNPxl creates precursor mass variants, submits data into the search engine, and summarizes the search results. (b) Results of data analysis procedure for a single dataset of yeast RNA-binding proteins. XIC, ID, and fractional mass filter excluded approximately two thirds (67%) of the overall 9,728 fragment spectra. Additionally, 16% of the spectra could be disregarded as these did not yield any database search result for a cross-linked heteroconjugate. Of the remaining 17% potential cross link candidates, 14% had a low score (E-value ≥ 0.01) yielding a final list of 317 (3%) potential cross link candidates for manual validation.