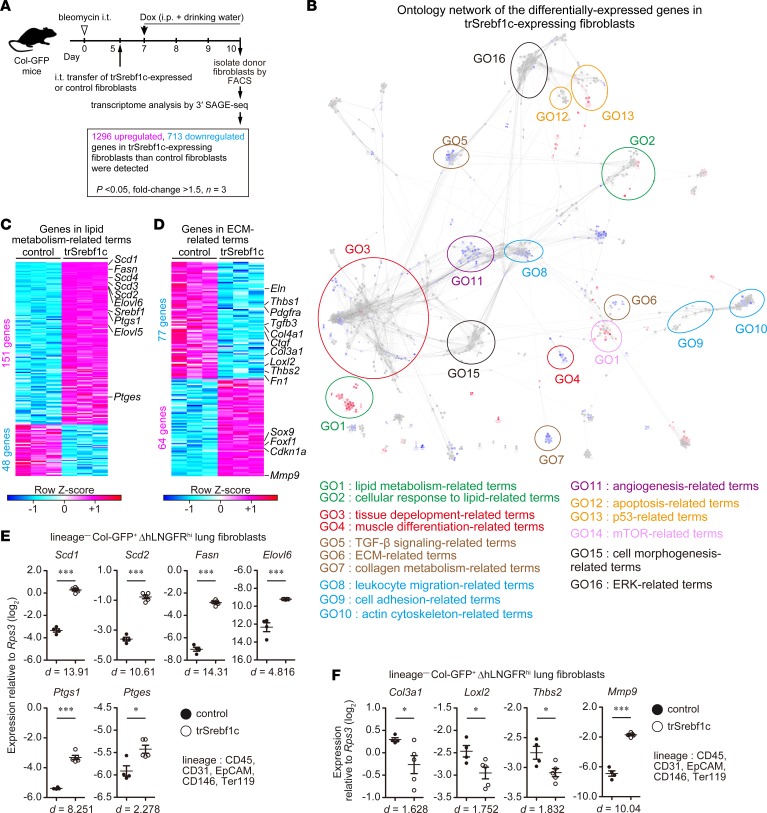
Figure 5. SREBP-1c broadly suppresses fibrosis-caused gene expression changes in activated lung fibroblasts.
(A) Experimental scheme of transcriptome analysis of intratracheally transferred genetically modified lung fibroblasts. In total, 2,009 genes were identified as differentially expressed genes as a result of ectopic expression of trSrebf1c. (B) Network of significantly enriched functional terms of the 2,009 genes. Biological events associated with the 2,009 genes were explored and term network was clustered using Cytoscape 3.3.0 with ClueGO and Allegrolayout plugins. Each node and its size represent functional term and enrichment significance, respectively. Red nodes represent functional terms for which genes from upregulated 1,296 genes comprised over 60% of all genes. Blue nodes represent functional terms for which genes from downregulated 713 genes comprised over 60% of all genes. Statistical significance was calculated for each term by using the 2-sided hypergeometric test with the Benjamini-Hochberg correction. (C and D) Details of lipid-related genes (C) and extracellular matrix–related (ECM-related) (D) genes identified by gene ontology network analysis. Each column represents group, whereas each row represents an individual gene. (E and F) qPCR analysis of expression changes of lipid-related (E) and ECM–related (F) genes. Graphs show the mean ± SEM of n = 4 (control) and n = 5 (trSrebf1c). A representative result of 2 independent experiments is shown. *P < 0.05, ***P < 0.001 (2-tailed unpaired Student’s t test). Effect size (d) is shown on the bottom of the graph. Col-GFP, Col1a2-GFP reporter; trSrebf1c active form of Srebf1c; ΔhLNGFR, truncated form of human low-affinity nerve growth factor receptor; SAGE, serial analysis of gene expression.