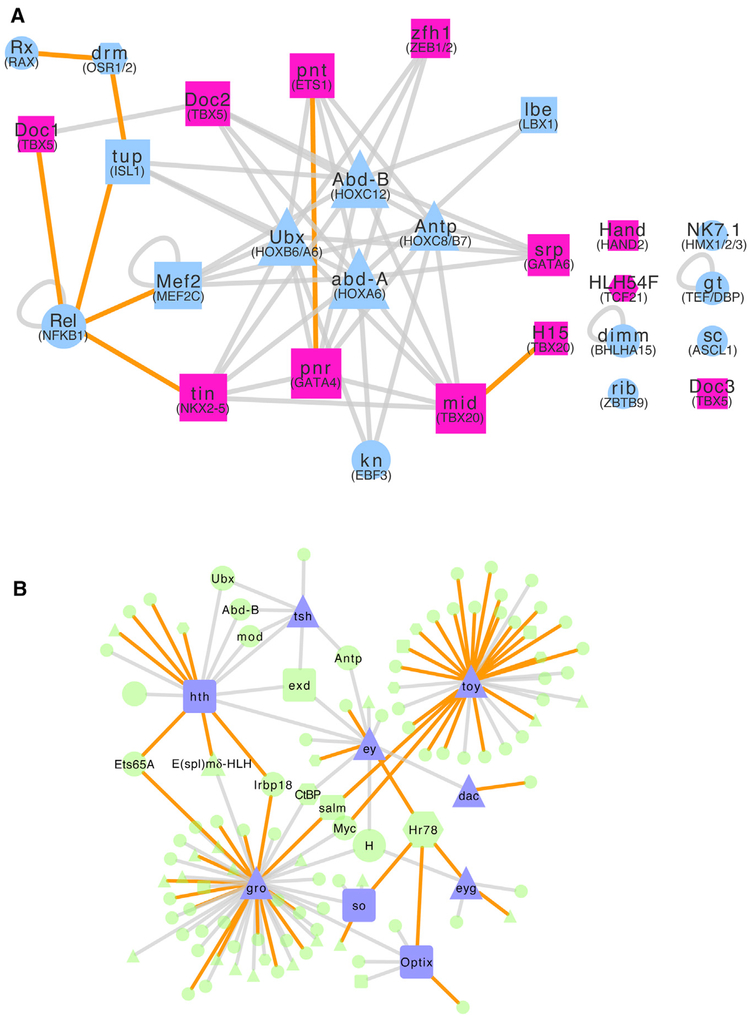
Figure 4. Integrated Fly TF-TF Interactome Identifies Candidate Human TFs Involved in Congenital Heart Disease and Eye Development.
(A) The Drosophila TF-TF interactome can be used to identify candidate heart development and CHD genes. Nodes: Drosophila TFs; edges: PPIs. Nodes with human orthologs that are annotated as CHD associated in HGMD (pink nodes) and their first neighbors (blue nodes) that are expressed in Drosophila heart tissue are shown. Unconnected components arose due to this tissue expression requirement. Square: the Drosophila gene and its human ortholog(s) are involved in CHD and heart development based on a literature search; triangle: the Drosophila gene is involved in heart development; hexagon: the Drosophila gene has a human ortholog involved in heart development; circle: neither the Drosophila gene nor human ortholog(s) are known to be involved in heart development. Node sizes roughly reflect node degrees. Orange edges indicate newly discovered interactions in this study. Human ortholog gene names are given in parentheses below the Drosophila gene name.
(B) The Drosophila TF-TF interactome can be used to identify candidate eye development genes. Nodes: Drosophila TFs; edges: PPIs. Nodes associated with eye development in Drosophila (based on Kumar, 2009; purple nodes) and their first neighbors (green nodes) are shown. Triangle: the Drosophila gene has eye development-related GO terms; hexagon: human, rat, mouse, or zebrafish (henceforth, vertebrate) ortholog of the Drosophila gene shown has eye development-related GO terms; square: the Drosophila gene (shown) and its vertebrate ortholog(s) have eye development-relatedGO terms; circle: neither the Drosophilagene nor vertebrate ortholog(s) have GO terms pertaining to eye development. Orange edges indicate newly discovered interactions in this study. Node sizes roughly reflect node degrees.