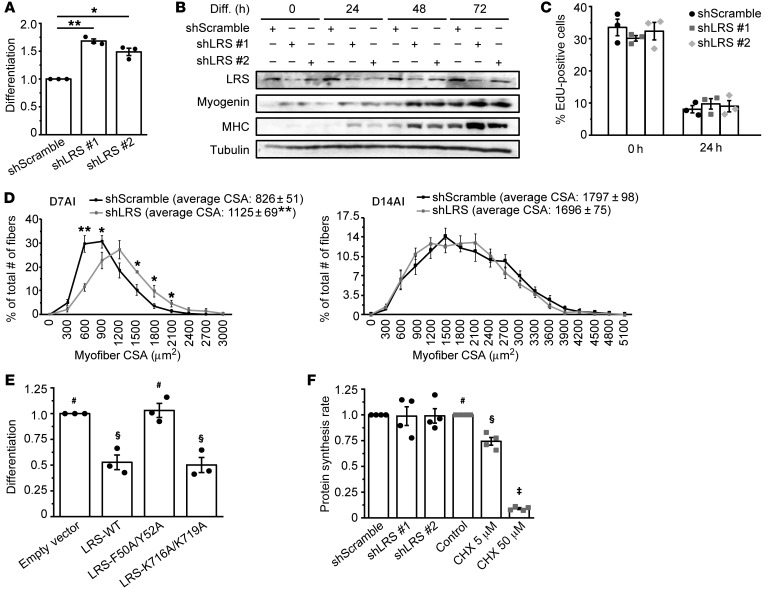
Figure 1. LRS negatively regulates myogenesis in a translation-independent manner.
(A) C2C12 cells were transduced with lentiviruses expressing shLRS or shScramble (control), puromycin-selected for 2 days, and differentiated for 72 hours, followed by measurement of differentiation index as described in the Supplemental Methods. Data were normalized to shScramble (n = 3). (B) Cells treated as in A were lysed every 24 hours and subjected to Western blotting analysis (n = 3). (C) Cells were treated as in A, and at 0 and 24 hours of differentiation, EdU incorporation was conducted for 2 hours as described in the Supplemental Methods (n = 3). (D) TA muscles were coinjected with BaCl2 and lentiviruses expressing shLRS or shScramble, and isolated on day 7 after injury (D7AI) (n = 5) and day 14 after injury (D14AI) (n = 6), followed by measurement of cross-sectional area (CSA) of regenerating myofibers as described in the Supplemental Methods. Data were presented as the size distribution of all myofibers with average CSA. (E) Cells were transfected with Myc-LRS-WT, F50A/Y52A, K716A/K719A, or empty vector (EV; control), selected with G418, and differentiated for 72 hours, followed by measurement of differentiation index. Data were normalized to EV (n = 3). (F) Cells were either transduced with lentiviruses expressing shLRS or shScramble, followed by puromycin-selection for 2 days (black bars), or treated with different concentrations of cycloheximide (CHX) for 24 hours (gray bars). Confluent cells were subjected to measurement of protein synthesis rate by [35S]Met/Cys metabolic labeling (n = 4). *P < 0.05, **P < 0.01 by 1-way ANOVA (A) or 2-tailed paired t test (D). The data in E and F denoted by #, §, ‡ are significantly different from each other by 1-way ANOVA (P < 0.05). All error bars represent SEM.