Fig. 1. TGF-β induces TAK1 isoform switch in epithelial and breast cancer cells.
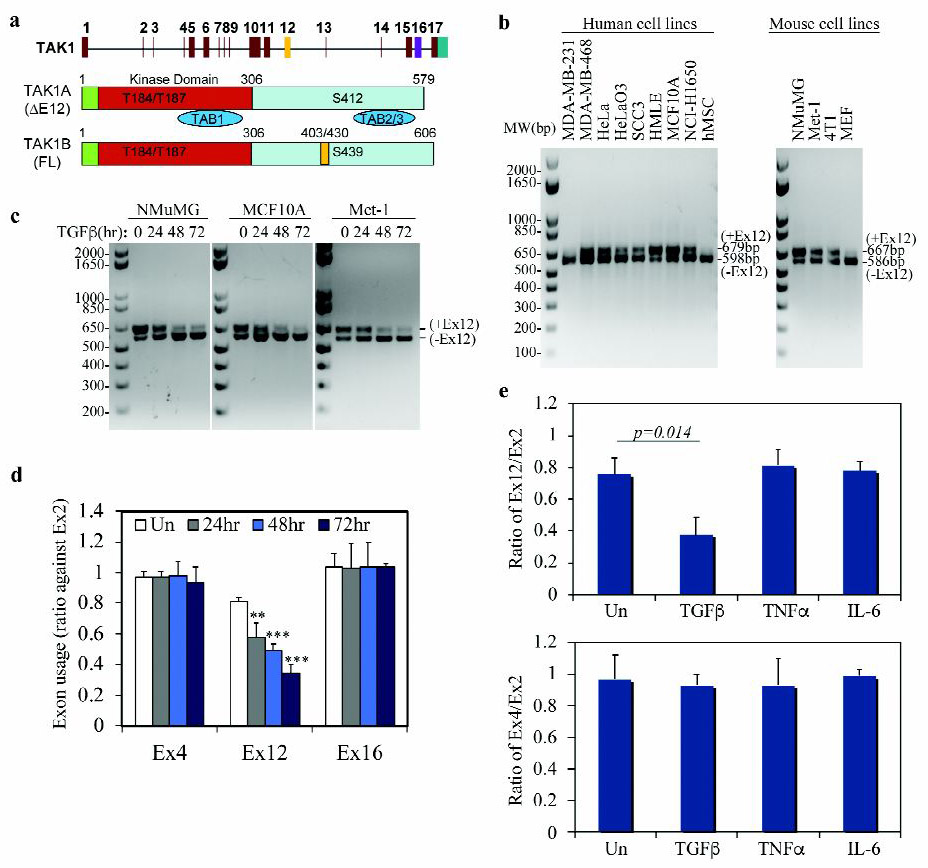
(a) Schematic diagram of structure of the TAK1 gene (top) and alternatively spliced mRNA transcripts of TAK1∆E12 with exon 12 exclusion and TAK1FL containing all exons.
(b) Detection of TAK1 isoforms with or without Exon 12 in various human and mouse cells using a RT-PCR reaction with one pair of primers.
(c) RT-PCR gel pictures showing that isoform switching of TAK1 in mammary epithelial NMuMG and MCF10A cells, and in breast cancer Met-1 cells treated with TGF-β.
(d) TGF-β specifically induces TAK1 exon 12 exclusion. qRT-PCR analysis of exon expression ratio of TAK1 variable exon 12 or 16 to standard exon 2 in NMuMG cells treated with TGF-β. Expression ratio of standard exon 4 to standard exon 2 was used as a control. Data are shown as mean ± SD (n=3), statistically significant difference between treated and untreated (Un) samples is indicated, **p<0.01, *** p<0.001.
(e) Exclusion of TAK1 exon 12 is specifically induced by TGF-β but not by TNFα or IL-6. qRT-PCR analysis of the exon ratio of TAK1 variable exon 12 to standard exon 2 (top), or exon 16 to standard exon 2 (bottom) in NMuMG cells treated with TGF-β, TNFα or IL6 for 72 h. Data are shown as mean ± SD (n=3), statistically significant difference between treated and untreated (Un) samples is indicated.
