Figure 7.
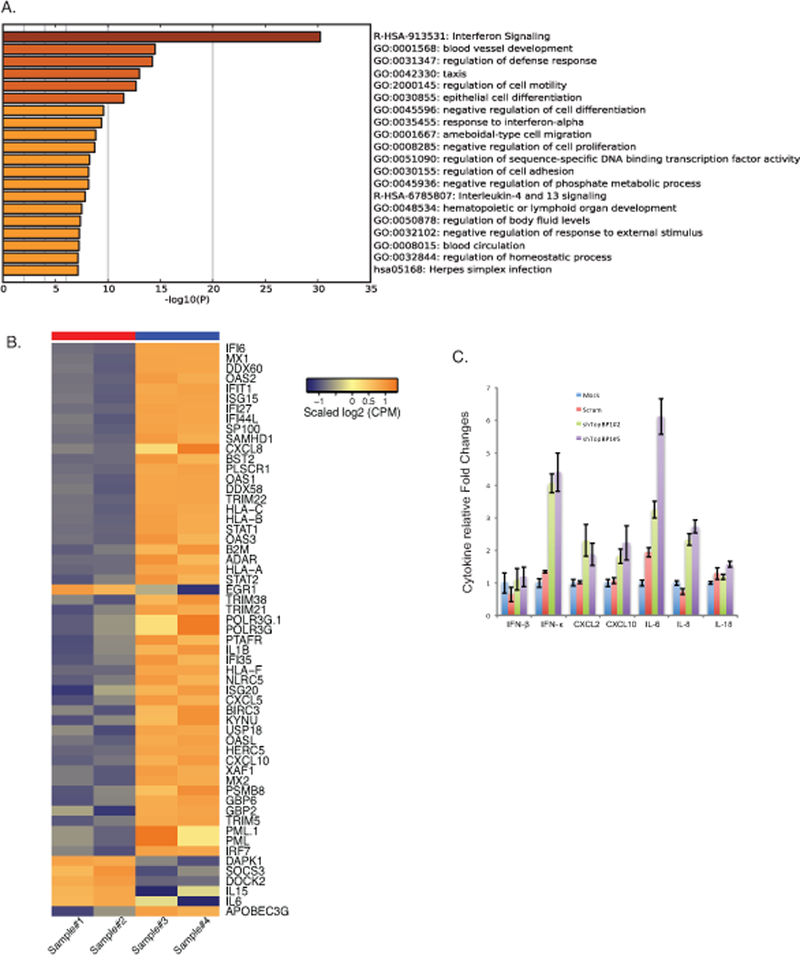
Downstream transcriptional targets of TopBP1 in HPV positive cells. A). RNA seq analysis was performed on CIN612 cells infected with scrambled shRNA control or shRNAs against TopBP1 following differentiation. Top 20 pathways regulated by TopBP1 was listed with Metascape online analysis tools. B). Heat maps of immune regulatory genes modulated by TopBP1 knockdown upon differentiation. Sample#1 and #2 are replicates of scramble CIN612 cells, while Sample #3 and #4 are replicates of CIN612 cells with TopBP1 knockdown. C) RT-PCR analysis of IFN-β, IFN-κ, CXCL2, CXCL10, IL-6, IL-8, and IL-18 in undifferentiated CIN612 cells with TopBP1 knockdown. GAPDH was used as internal control and for normalization of the data. Data=mean +/− standard error. p-value <0.05. All results are representative of observations from 2 or more independent experiments.
