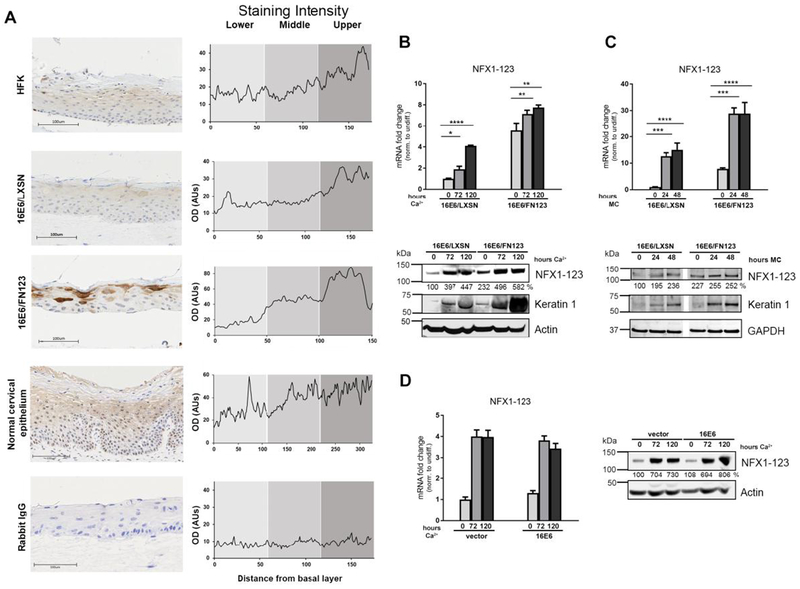
Fig 1. NFX1-123 expression increases with keratinocyte differentiation.
(A) Organotypic raft cultures were grown using HFKs, HFKs serially transduced with 16E6 and vector control (16E6/LXSN), and HFKs transduced with 16E6 and FLAG-tagged NFX1-123 overexpression construct (16E6/FN123). These rafts and normal cervical epithelial biopsies were stained for NFX1-123 protein via immunohistochemistry or stained using a rabbit polyclonal IgG as a control. Staining intensity was measured over a linear distance from the basal layer in ImageJ and the average of four plots over two sections is shown. (B and C) 16E6/LXSN and 16E6/FN123 HFKs were differentiated with 1.8mM Ca2+ treatment for 0, 72, or 120 hours (B) or suspension in 1.7% methylcellulose medium (MC) (C) and total mRNA and protein collected. Mean expression of NFX1-123 mRNA was measured by qPCR and compared to 16E6/LXSN 0 hours. Protein levels of NFX1-123 were measured by Western blot using actin or GAPDH as a loading control. Induction of the differentiation marker Keratin 1 indicates successful differentiation. White spaces in Western blots indicate removal of empty or irrelevant lanes from original image for clarity. (D) HFKs transduced with empty vector or with 16E6 were differentiated with 1.8mM Ca2+ treatment for 0, 72, or 120 hours, and total mRNA and protein collected. Mean expression of NFX1-123 mRNA was measured by qPCR and each sample was compared to HFK/vector 0 hours. Protein levels of NFX1-123 were measured by Western blot using actin as a loading control. For B, C, and D, mRNA expression of NFX1-123 for each sample was normalized to the housekeeping gene 36B4, and all error bars represent 95% confidence intervals from replicates. Protein expression of NFX1-123 for each sample was quantified using ImageJ and normalized to the loading control for that sample. One representative experiment is shown from at least three conducted in biologically independent HFK cell lines. Statistical significance was calculated using one-way ANOVA with Bonferroni correction. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001