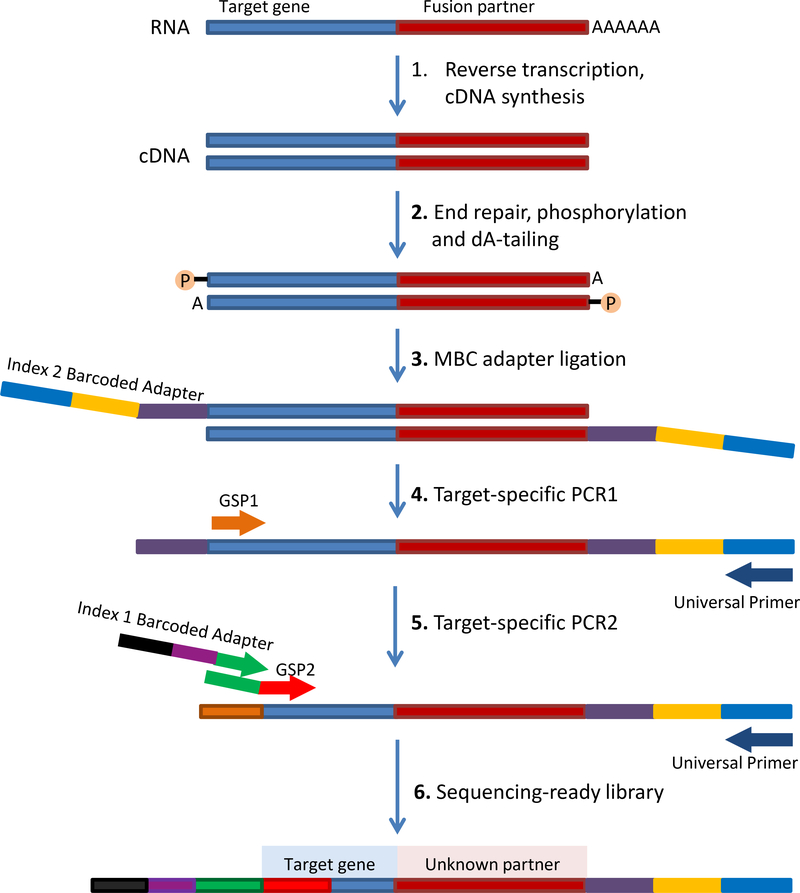
Figure 1.
Schematic of Archer’s Anchored Multiplex PCR (AMP™) workflow. Adapted from www.archerdx.com. RNA is extracted from tumor formalin-fixed paraffin-embedded material followed by cDNA synthesis. cDNA undergoes end repair, dA tailing and ligation with half-functional Illumina molecular barcode adapters (MBC). Cleaned ligated fragments are subject to two consecutive rounds of PCR amplifications using two sets of gene specific primers (GSP1 pool used in PCR1 and a nested GSP2 pool designed 3’ downstream of GSP1 and used in PCR2) and primers complementary to the Illumina adapters. At the end of the two PCR steps, the final targeted amplicons are ready for 2×150bp sequencing on an Illumina MiSeq sequencer.