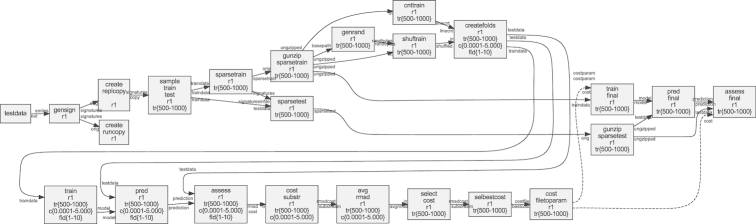
Figure 5.
Directed graph of the machine learning in drug discovery case study workflow, plotted with SciPipe’s workflow plotting function. The graph has been modified for clarity by collapsing the individual branches of the parameter sweeps and cross-validation fold generation. The layout has also been manually made more compact to be viewable in print. The collapsed branches are indicated by intervals in the box labels. tr{500-8000} represent branching into training data set sizes 500, 1,000, 2,000, 4,000, 8,000. c{0.0001-5.0000} represent cost values 0.0001, 0.0005, 0.001, 0.005, 0.01, 0.05, 0.1, 0.25, 0.5, 0.75, 1, 2, 3, 4, and 5, while fld{1-10} represent cross-validation folds 1−10. Nodes represent processes, while edges represent data dependencies. The labels on the edge heads and tails represent ports. Solid lines represent data dependencies via files, while dashed lines represent data dependencies via parameters, which are not persisted to file, only transmitted via RAM.