Abstract
In mammalian ventricular cardiomyocytes, invaginations of the surface membrane form the transverse tubular system (T-system), which consists of transverse tubules (TTs) that align with sarcomeres and Z-lines as well as longitudinal tubules (LTs) that are present between Z-lines in some species. In many cardiac disease etiologies, the T-system is perturbed, which is believed to promote spatially heterogeneous, dyssynchronous Ca2+ release and inefficient contraction. In general, T-system characterization approaches have been directed primarily at isolated cells and do not detect subcellular T-system heterogeneity. Here, we present MatchedMyo, a matched-filter-based algorithm for subcellular T-system characterization in isolated cardiomyocytes and millimeter-scale myocardial sections. The algorithm utilizes “filters” representative of TTs, LTs, and T-system absence. Application of the algorithm to cardiomyocytes isolated from rat disease models of myocardial infarction (MI), dilated cardiomyopathy induced via aortic banding, and sham surgery confirmed and quantified heterogeneous T-system structure and remodeling. Cardiomyocytes from post-MI hearts exhibited increasing T-system disarray as proximity to the infarct increased. We found significant (p < 0.05, Welch’s t-test) increases in LT density within cardiomyocytes proximal to the infarct (12 ± 3%, data reported as mean ± SD, n = 3) versus sham (4 ± 2%, n = 5), but not distal to the infarct (7 ± 1%, n = 3). The algorithm also detected decreases in TTs within 5° of the myocyte minor axis for isolated aortic banding (36 ± 9%, n = 3) and MI cardiomyocytes located intermediate (37 4%, n = 3) and proximal (34 ± 4%, n = 3) to the infarct versus sham (57 ± 12%, n = 5). Application of bootstrapping to rabbit MI tissue revealed distal sections comprised 18.9 ± 1.0% TTs, whereas proximal sections comprised 10.1 ± 0.8% TTs (p < 0.05), a 46.6% decrease. The matched-filter approach therefore provides a robust and scalable technique for T-system characterization from isolated cells through millimeter-scale myocardial sections.
Introduction
In ventricular cardiomyocytes, Ca2+-induced-Ca2+ release begins with the propagation of an action potential along the cell membrane, eliciting sarcolemmal Ca2+ influx via channels (LCCs). These channels are primarily localized within sarcolemma (SL) invaginations comprising the tubular system (T-system) (1). Localized Ca2+ entry via LCCs triggers neighboring ryanodine receptors located in the sarcoplasmic reticulum (SR) membrane to open and release Ca2+ from SR stores (1, 2, 3, 4). The spatiotemporal homogeneity of Ca2+-induced-Ca2+ release is critical in maintaining efficacious contraction of the heart. In rabbit, canine, and human ventricular cardiomyocytes, this transverse T-system is comprised predominantly of transverse tubules (TTs) coinciding with the Z-lines. In rodent ventricular cardiomyocytes, the T-system includes both TTs as well as significant longitudinal tubule (LT) or axial tubule components (5, 6). In failing hearts, severe morphological changes in the T-system of ventricular cardiomyocytes promote impairment of critical excitation-contraction-coupling Ca2+-handling mechanisms (1, 7, 8) that drive Ca2+ dysregulation and cardiac dysfunction (9, 10, 11).
Advances in fluorescence-based microscopy offer outstanding potential to probe mechanisms governing T-system remodeling. However, the nature of T-system remodeling is diverse, with some pathologies almost strictly presenting tubule absence (TA), whereas others can exhibit tubules in nontransverse arrangements. Among the disease models that manifest evident changes in the cardiomyocyte T-system, distinctive patterns have emerged, from reduced TT density in type II diabetic mice (12) to the coupled observation of TA and increased LT density with differing patterns for congenital heart failure (13), spontaneous hypertension/heart failure (14), and other disorders (15). Sub-micron-resolution confocal microscopic images exist for several models of heart failure (concentric hypertrophy and dilated hypertrophy, for instance) in species including mouse (10, 13, 16, 17, 18), rabbit (19, 20, 21), canine (22, 23), and human (24, 25). However, quantitative assessments of subcellular T-system remodeling have been limited.
Characterization of images and correlating structural features with cardiac function (26) represent significant challenges that permit computational solutions. Several methods were introduced to automate the characterization of cardiomyocyte T-system. Among these approaches is the “TT index” or “TTPower,” whereby the relative spacing and angle of TT arrangements are estimated by Fourier analysis (27, 28, 29). TTPower correlates well with the severity of remodeling but provides limited structural classification. More recent methods apply some level of preprocessing to increase the amplitude of the signal arising from the TTs (27, 28). The most common metric appears to be binary thresholding, which increases contrast by converting all pixels to binary values depending on their value relative to a user-determined threshold. Others utilize morphological transformations, which produce “skeletonized” representations of the T-system that enhance the TT index (27, 28, 30). Recent uses of techniques such as histogram equalization (31) have been tuned to improve existing analysis approaches (27, 28, 30). The advantage of these approaches is that they assume little prior knowledge about the structure of the T-system, yet their applications have been limited to single-cardiomyocyte preparations.
Here, we present MatchedMyo, a matched-filter based image processing algorithm for the subcellular characterization of the cardiomyocyte T-system. The method relies on images, or filters, representing features of interest such as TTs, LTs, and TA. In contrast to methods that binarize images to highlight features, this algorithm preserves the dynamic range of the original image. We outline the corresponding workflow in Fig. 1, which we use to characterize the total cellular content of intact TTs, LTs, and TA in isolated sham, myocardial infarction (MI), and dilated cardiomyopathy via ascending aortic banding (AB) cells as well as MI tissue sections (cell and tissue preparation summarized in Supporting Materials and Methods, Section S3.1).
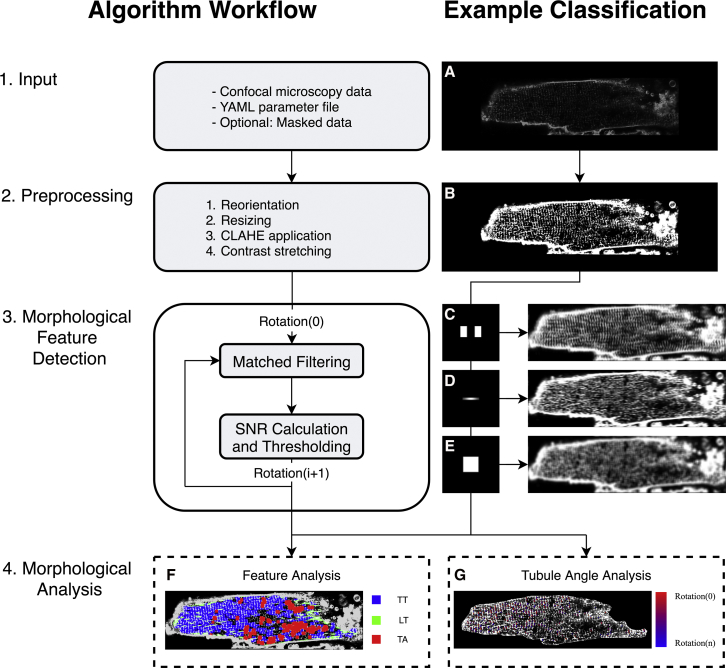
Figure 1.
Unicellular matched-filtering protocol: 1) input of confocal microscopy data, YAML (.yml) parameter file, and optional user generated mask for that data, for which (A) shows sample input image. YAML parameter file construction is explained in Supporting Materials and Methods, Section S1.4; 2) integrated preprocessing routines in which (B) shows sample preprocessed image; 3) morphological feature detection via matched filtering and signal/noise ratio calculation. (C–E) Matched filters and sample signal/noise ratio calculation via convolution for TT, LT, and TA filters, respectively; 4) morphological analysis of features, feature density, and feature rotation. (F) Sample morphological feature content analysis exhibiting high TT content relative to other morphological features. (G) Sample tubule striation angle analysis with high angle variance. To see this figure in color, go online.
Materials and Methods
Matched-filter theory and application
We developed a computer program that analyzes microscopic images of cardiac cells and measures where structures involved in calcium handling are located. This is important because changes in cell calcium can contribute to disease. Our program was based on techniques from signal-processing theory and used an approach called “matched filtering.” In essence, our matched-filtering approach scans cardiomyocytes for regions that resemble a representative image, or filter, of TT and LT features. This scanning is accomplished by a mathematical operation, convolution, of the cardiomyocyte image with the TT filter. The result of this convolution is a new image, for which each pixel represents the propensity of finding the filter at the corresponding location in the cardiomyocyte. We utilize a bank of rotated TT filters to tolerate moderate variations in TT orientation, followed by automated, postprocessing techniques to classify detected features.
More formally, matched filtering determines whether a known signal s, or in vector form, , is embedded in statistically uncorrelated noise, , via the relationship
| (1) |
where y is the likelihood the known signal is embedded in the measured signal, is the measured signal (32), and Cn is the noise covariance (, where signifies the expectation of a random, spatially distributed noise vector, ). The overarching principle in matched filtering is to identify a matched filter, , that maximizes the signal/noise ratio for a measurement, (32), via
| (2) |
where denotes the filter’s transpose. We utilize this approach in the first stage of our workflow to assess the propensity that a filter representing a feature of interest is present in a given data set. Often the signal or multiple instances thereof is embedded within a larger data set (), such as an image, in which case determining the location of within is commonly performed by convolving the filter with the image, , via
| (3) |
| (4) |
where Y(x, y) is the propensity of finding the known signal, , in the image, , at (x, y), whereas and represent the discrete Fourier transform and inverse Fourier transform, respectively, to solve Eq. 3 via the convolution theorem. Probable detections of the filter within Yi are then based on identifying positions at which the response is above a user-specified threshold criterion, λi. To classify where cellular microstructure resembles known filters, we evaluate
| (5) |
The optimal filter is chosen as max({Y1, Y2, …}), so long as there exists at least one Yi for which Yi(x, y) > λi. Regions of the test confocal images that returned responses below the threshold parameters for all filters considered are designated as “uncharacterized.” Matched-filter data reported herein for TT, LT, and TA filters are reported as percent characterized cell area of total cell area for each filter.
Detection by convolution
Beyond classifying whether cellular microstructure resembles known filters, our workflow is tasked with designating regions of T-system absence as well as unclassified TT structure. For this reason, we present the following detection scheme. We assess the likelihood of subcellular content via
| (6) |
| (7) |
where ∗ represents the convolution of the filter, , , and , with the confocal microscopy image, , and λi represents the user-determined threshold. A region is considered uncharacterized if no is indicated as a hit via this criterion.
Filter bank
In our approach, we additionally consider filter rotations to detect alternate tubule orientations through defining a bank of filter rotations for each matched filter spaced at 5° increments. The optimal rotation for a given filter is determined by evaluating each rotated filter with the data; the rotated filter that generates the highest above-threshold response is elected as the most likely orientation of a given feature, j:
| (8) |
| (9) |
where R(ϕj) represents the rotation of filter h by ϕj.
Matched-filter determination
Here, we determine the matched filters based on manually identifying representative T-system structure in the confocal data. The three morphological characteristics that we identified were TT, LT, and TA regions. The filters are shown in Fig. 1. The TT filter consisted of two separated binary TT structural elements. The complement to this filter was utilized as a penalty filter as described in Supporting Materials and Methods, Section S2.1. The LT filter consisted of a 1-sarcomere-wide longitudinal structural element with varying intensity according to a normal distribution along the x axis. This allowed for greater specificity of LT versus TT detection by attenuating the spectral overlap of the filters. The TA filter was a sarcomere-length square smoothing filter. The presence of subthreshold pixels in the smoothed image indicated the presence of sarcomere-sized regions of low signal density. Details for selecting optimal threshold parameters and corresponding receiver operator characteristic curves are provided in Supporting Materials and Methods, Section S4.1.
Analysis of TTs within 5° of myocyte minor axis
It has been shown that TTs are not strictly aligned perpendicular to the SL in healthy myocytes during rest (33), and the associated sarcomeric cytoskeleton can be perturbed in disease states (34, 35). Because of this, we were interested in quantifying tubule orientation across the etiologies considered herein. Because our method relies on convolution, quantification of local tubule orientation, termed tubule striation angle, is straightforward. Isolated cardiomyocytes and tissue sections were manually oriented via the included graphical user interface such that the observable majority of TTs were parallel to the y axis. We manually oriented the cardiomyocytes for simplicity, but in principle, this process could be automated by utilizing angles inferred from the power spectral density of the low spatial frequency information of the outer SL. Filters were rotated at five increments, between −25 and 25, and convolved with the image. TT filter hits at −5, 0, and 5, relative to the specified minor axis, were compared to the total number of hits across all rotations.
Tissue section analysis
Relative to analysis of isolated cardiomyocytes, the tissue-level characterization presented unique challenges, including segmentation of nonmyocyte regions. To address this challenge, we introduced a penalty filter to increase specificity of the TT filter in the presence of noncardiac regions (see Supporting Materials and Methods, Section S2.1 for more details). Thus, for these analyses, we did not mask the cell boundaries as was done for the isolated cardiomyocytes. Given that no mask was applied to the images, we found that the performance of the LT filter and tubule striation angle analysis was substantially degraded and thus was not included.
Computational tools
All aforementioned numerical procedures were conducted using the Python 2.7 libraries NumPy, SciPy, and OpenCV-Python. Data processing was performed using SciPy and Jupyter Notebooks. All source code is provided at https://bitbucket.org/pkhlab/matchedmyo. Issue tracking and feature requests can be posted at https://bitbucket.org/pkhlab/matchedmyo/issues. A limited set of tests can be performed with the code at http://athena.as.uky.edu/node/7; however, for figure-quality images and custom analyses, download of the MatchedMyo software is strongly recommended. Details of the statistical analyses, algorithm use, installation, and algorithm refinements are provided in the Supporting Materials and Methods. We have provided detailed installation and operation instructions; see Supporting Materials and Methods, Section S1.
Results and Discussion
Analysis of isolated cardiomyocytes
Cardiomyocytes from sham rat model
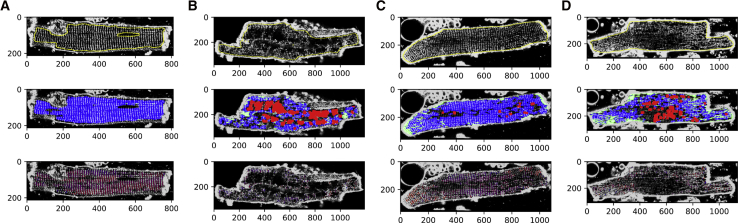
We first demonstrate the matched-filtering approach using images of sham cells (Fig. 2 A), in which the TTs exhibit a regular, striated pattern (see Supporting Materials and Methods, Section S3.1 for details on cell isolation and imaging). Nevertheless, subregions of the cardiomyocyte presented an appreciable degree of spatial and angular variation. In the middle row of Fig. 2 A, we present the classification results, for which the TT, LT, and TA responses are overlaid as blue, green, and red, respectively. To improve the classification results, we utilized a mask to exclude the cell membrane and nuclei in the field of view. In this image, the TT structure predominates over the LT and TA signals (middle row of Fig. 2 A), which we summarize in Table 1. Finally, we present in the bottom row of Fig. 2 A a heatmap of tubule striation angle superimposed onto the original image, in which red corresponds to a TT filter hit at −25° and blue corresponds to a TT filter hit at +25°.
Figure 2.
Algorithm application to isolated cardiomyocytes. Cardiomyocytes from (A) sham, (B) dilated cardiomyopathy via ascending AB, (C) MI (distal to infarct), and (D) MI (proximal to infarct) models are shown. (Top row) Preprocessed confocal images with outlines of image masks marked in yellow are shown. (Middle row) MatchedMyo output with transverse tubule (TT), LT, and TA filter hits marked in blue, green, and red, respectively, are shown. (Bottom row) A heatmap of tubule striation angle is shown superimposed onto the image, where red corresponds to TT hits at −25° and blue corresponds to TT hits at 25°. To see this figure in color, go online.
Table 1.
Quantification of Content Present within Myocytes from Differing Disease Etiologies, Presented as Percentage of Cell Area
| Case (n Cells) | TT (%) | LT (%) | TA (%) | TT ± 5° Minor Axis (%) |
|---|---|---|---|---|
| Sham (5) | 78 ± 11 | 4 ± 2 | 9 ± 5 | 57 ± 12 |
| AB (3) | 19 ± 14a | 5 ± 1 | 37 ± 13 | 36 ± 9a |
| MIDistal (3) | 56 ± 15 | 7 ± 1 | 11 ± 6 | 48 ± 6 |
| MIIntermediate (3) | 57 ± 4a | 10 ± 2a | 10 ± 7 | 37 ± 4a |
| MIProximal (3) | 55 ± 16 | 12 ± 3a | 12 ± 6 | 34 ± 4a |
| Distal tissue (1000) | 18.9 ± 1.0 | – | 9.5 ± 2.0 | – |
| Proximal tissue (1000) | 10.1 ± 0.8b | – | 16.0 ± 2.0 | – |
Refer to the Supporting Material for full range of cardiomyocytes considered herein. Data presented as mean SD. TT denotes a “hit” from the transverse tubule filter. Similarly, LT and TA denote “hits” from the longitudinal and tubule absence filters, respectively. Distal and Proximal tissue results were analyzed using the bootstrapping method explained in Supporting Materials and Methods, Section S3.4; n denotes number of synthetic images created for analysis.
p < 0.05 compared to sham result.
p < 0.05 compared to distal result.
In principle, similar conclusions can be drawn by visual inspection of these images or application of other analyses, such as the TTPower method (see Supporting Materials and Methods, Section S4.3 for a comparison to Fourier transform TT density estimation). However, one benefit of the matched-filtering approach is the ability to detect spatially heterogeneous variations in the T-system. For instance, the TT striations are not strictly straight throughout the cardiomyocyte interior. Rather, the striations appear to accommodate roughly perpendicular orientations relative to the outer SL along the major axis. In traditional TT index or power methods, such spatial variations in angular frequencies would present a diffuse signal response in the Fourier domain, leading to a potential misclassification of subcellular structure, unless the outer SL is masked or the region of interest is restricted to a small area. Hence, our matched-filtering approach is fundamentally different from techniques such as AutoTT (28) because our method computationally searches for arbitrary features within images versus specifically characterizing TT periodicity or morphology. Further, because our method relies on convolution, we were able to quantify the angular variation (Table 1). Here, we find that sham cells present 57 ± 12% of striations within 5° of the myocyte minor axis (see Supporting Materials and Methods, Section S3.4 for details on statistical analysis). We also note that in rodent species, the T-system presents considerable branching that manifests in significant axial contributions (5). These features were not evident in the cells considered here, owing most likely to the lack of confocal z-stacks. For the LT and TA criterion, we report no significant responses in Fig. 2 A.
Remodeling in AB cardiomyocytes
We next investigated T-system remodeling in tissue derived from a rat model of dilated cardiomyopathy based on AB (36). In these animals, AB emulates pressure overload, which is known to drive hypertrophic remodeling broadly throughout cardiac tissue (37). Commonly, tissue derived from AB mice present considerable TA, which correlates with impaired contractile function (29, 38). Here, we applied the algorithm to the tissue shown in Fig. 2 B, using identical filters and thresholds from the previous sham analysis. As shown in Fig. 2 B and Table 1, we find that TT content is significantly reduced (p < 0.05) in AB isolated cardiomyocytes (19 ± 14%) relative to sham (78 ± 11%). This results in pervasive TA regions for AB cardiomyocytes (37 ± 13%) versus sham (9 ± 5%). TA regions appear to be localized to the midline of the cell interior, whereas intact structure is mostly maintained adjacent to the outer plasma membrane. Although TTs appear to be adjacent to the outer SL, there remains a significant (p < 0.05) decrease in TTs within 5° of the myocyte minor axis (36 ± 9%) versus sham. We report no significant changes in LT content for AB cardiomyocytes (5 ± 1%) versus sham (4 ± 2%).
Cardiomyocytes in MI rat heart
MI introduces a localized necrotic region in tissue. Cells adjacent to this necrotic region and border zone exhibit varying degrees of remodeling (10). Here, we applied the algorithm to cardiomyocytes isolated from regions distal, intermediate, and proximal to an MI, shown in Fig. 2, C and D (medial cardiomyocytes not shown in figure; see Fig. S4 for all MI cardiomyocytes considered herein). These cells were obtained from an infarcted heart after explantation, for which the border zone was removed and the remaining noninfarcted regions of the heart were divided into three equal-sized regions as described previously (39). Relative to the sham cardiomyocytes, proximal cardiomyocytes exhibited a substantial increase in LTs (12 ± 3%, p < 0.05) as well as a decrease in TT density, indicated by TA filter hits (12 ± 6%, Table 1). Cardiomyocytes from the intermediate region presented both LTs (10 ± 2%, p < 0.05) and TA (10 ± 7%), although to a lesser extent than the proximal cells.
Finally, we confirm for distally derived tissue that for the four metrics used in this study—TT, LT, TA, and tubule striation angle—the differences observed were insignificant compared to sham cardiomyocytes. Relative to isolated cardiomyocytes from AB models, we see a substantial increase in LTs and a decreased amount of TA in MI. As with AB cardiomyocytes, a significant (p < 0.05) decrease in the amount of TTs present within 5° of the myocyte minor axis was observed in proximal tissue (34 ± 4%) and intermediate tissue (37 ± 4%) versus sham. The main implication of this finding is that remodeling after MI appears to give rise to LTs and TT reorganization that is distinct from TA patterns exhibited in pressure overload models while exhibiting similar decreases in TTs within 5° of the myocyte minor axis. Further, the extent of remodeling parallels distance from MI, whereas TT density appears to have an inverse relationship with proximity to the MI.
Analysis of cardiac tissue from infarcted rabbit heart
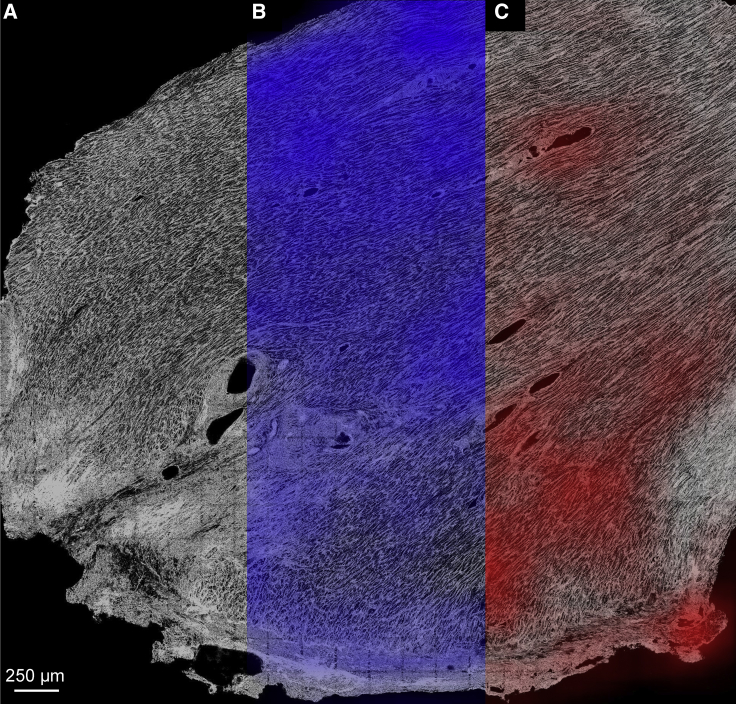
In Fig. 3, we present results for application of the MatchedMyo algorithm to MI tissue, which provides detail of the local tissue environment and the spatial distribution of tissue remodeling. Fig. 3, A–C show portions of the infarcted tissue with no filtering, TT filtering, and TA filtering, respectively. Fig. 3, B and C show heatmaps of TT and TA filter hits superimposed onto the tissue image. As observed previously in Fig. 2, C and D, there was a marked decrease in the TT content, paralleled by an increase in TA detection, as the proximity to an infarct increased. Relative to the distal subsection, the proximal subsection (shown in further detail in Fig. S7, C and D, respectively) presented, on average, 53.4% detection with the TT filter across the same area (see Table 1). This is paralleled by an increase in TA detection, with the proximal subsection presenting 68.4% more TA content than the distal subsection on average (see Supporting Materials and Methods, Section S3.4 for further details on statistical analysis).
Figure 3.
Application of TT filter (blue) and TA filter (red) to 3.9 × 4.1 mm2 sections of infarcted rabbit tissue where infarct is located at the bottom of the image, indicated by severe fibrosis. (A) A raw confocal tile scan exhibiting varying degrees of T-system remodeling is shown. (B and C) Smoothed TT and TA filter detections are shown with heatmaps normalized to maximal density of filter detections. A threshold was applied to the smoothed TA hits to exclude blood vessels. Striations arise from close apposition of punctate TT patterns typically observed in rabbit cardiomyocytes (43). As summarized in Table 1, the distal region presented 18.9 ± 1.0% TT detection compared to 10.1 ± 0.8% in the proximal region. TA detection paralleled TT detection, with the distal region exhibiting 9.5 ± 2.0% and the proximal region exhibiting 16.0 ± 2.0% TA detection. To see this figure in color, go online.
Limitations
A limitation of the software is the classification of organelles as TA because of their lack of fluorescence. This can be addressed by applying a DAPI stain to the cardiomyocyte and thresholding the image to create a mask. Instructions for mask application are shown in Supporting Materials and Methods, Section S1.4.
Conclusions
Our matched-filtering-based technique complements existing computational algorithms for performing automated analyses of cardiomyocyte T-system (27, 28). This approach leverages detailed structural information for the features of interest, based on kernels reflective of TT, LT, and TA morphology. Its reliance on convolution permits localized assessments of remodeling that scale to tissue-level preparations. Evidently the approach performed remarkably well across cardiomyocytes from different species, disease etiologies, and fields of view. Our results suggest that the matched-filtering approach performs favorably for detecting different extents and modalities for T-system configurations in confocal microscopic images and reveal patterns of T-system structure unique to sham, MI, and AB cardiomyocytes. We found that the approach is robust to regional variation in subcellular structure and noise while providing, to our knowledge, a first-of-its-kind method scalable to tissue-level imaging.
For the cardiomyocytes we considered, the approach supported prior work revealing interesting trends in regional variation of cardiomyocyte T-system, such as the heterogeneity of TT organization in sham cells, as well as intracellular remodeling that depended on the distance from MI (39, 40). We anticipate that these variations may give important clues to how cells adapt to their environment. This topic was previously explored by Guo et al. (11), who cataloged diverse morphological configurations of the cardiomyocyte T-system with respect to species and disease etiology.
Correlations between T-system disorganization and Ca2+ release dyssynchrony in numerous models of cardiac dysfunction (10, 13, 41, 42) suggest strong linkages between cardiomyocyte remodeling and pathophysiological function. It has been observed that SL proteins can redistribute under certain conditions that correlate with Ca2+ mismanagement, with no observation of increased TA (13). Accordingly, it will be invaluable to investigate the potential correlations between T-system remodeling and protein localization, as well as the extent to which those relationships influence normal physiological function. In this regard, although the data considered in this study were limited to confocal images of dye-labeled SL, it is straightforward to adapt the algorithm to diverse data sets. Hence, in the event that filters representing features of interest can be identified, the approach can be adapted to a host of different labeling and imaging techniques, as well as cells isolated from a spectrum of tissue types.
Author Contributions
D.F.C.: algorithm design, application/optimization of algorithm, wrote manuscript. S.R.B.: algorithm design, edited manuscript. A.C.S. imaged tissue and edited manuscript. F.B.S. provided tools for tissue data acquisition and edited manuscript. M.F. imaged isolated myocytes. W.E.L. provided tools for isolated myocyte data acquisition and edited manuscript. P.M.K.-H.: research design, algorithm design, wrote manuscript.
Acknowledgments
We thank Noah Adler for his support in integrating our utilities onto a web platform and Darin Vaughan and Ken Campbell for critical discussion of the manuscript, as well as Dr. Thomas McCoy for bootstrapping technique advice.
This work was supported by the Maximizing Investigators’ Research Award (R35) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number R35GM124977, as well as an Institutional Development Award from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103527. D.F.C. would also like to acknowledge support through the American Heart Association grant 17UFEL33490002.
Editor: Mark Cannell.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.03.010.
Supporting Material
References
- 1.Bers D.M. Volume 1. Kluwer Academic Publishers; Norwell, MA: 2001. p. 427. (Excitation-Contraction Coupling and Cardiac Contractile Force.). [Google Scholar]
- 2.Litwin S.E., Li J., Bridge J.H. Na-Ca exchange and the trigger for sarcoplasmic reticulum Ca release: studies in adult rabbit ventricular myocytes. Biophys. J. 1998;75:359–371. doi: 10.1016/S0006-3495(98)77520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern M.D. Theory of excitation-contraction coupling in cardiac muscle. Biophys. J. 1992;63:497–517. doi: 10.1016/S0006-3495(92)81615-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X.H., Protasi F., Franzini-Armstrong C. Molecular architecture of membranes involved in excitation-contraction coupling of cardiac muscle. J. Cell Biol. 1995;129:659–671. doi: 10.1083/jcb.129.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soeller C., Cannell M.B. Examination of the transverse tubular system in living cardiac rat myocytes by 2-photon microscopy and digital image-processing techniques. Circ. Res. 1999;84:266–275. doi: 10.1161/01.res.84.3.266. [DOI] [PubMed] [Google Scholar]
- 6.Amsellem J., Delorme R., Ojeda C. Transverse-axial tubular system in Guinea pig ventricular cardiomyocyte: 3D reconstruction, quantification and its possible role in K+ accumulation-depletion phenomenon in single cells. Biol. Cell. 1995;85:43–54. [PubMed] [Google Scholar]
- 7.Bers D.M. Altered cardiac myocyte Ca regulation in heart failure. Physiology (Bethesda) 2006;21:380–387. doi: 10.1152/physiol.00019.2006. [DOI] [PubMed] [Google Scholar]
- 8.Pinali C., Bennett H., Kitmitto A. Three-dimensional reconstruction of cardiac sarcoplasmic reticulum reveals a continuous network linking transverse-tubules: this organization is perturbed in heart failure. Circ. Res. 2013;113:1219–1230. doi: 10.1161/CIRCRESAHA.113.301348. [DOI] [PubMed] [Google Scholar]
- 9.Louch W.E., Sejersted O.M., Swift F. There goes the neighborhood: pathological alterations in T-tubule morphology and consequences for cardiomyocyte Ca2+ handling. J. Biomed. Biotechnol. 2010;2010:503906. doi: 10.1155/2010/503906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louch W.E., Mørk H.K., Sejersted O.M. T-tubule disorganization and reduced synchrony of Ca2+ release in murine cardiomyocytes following myocardial infarction. J. Physiol. 2006;574:519–533. doi: 10.1113/jphysiol.2006.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo A., Zhang C., Song L.S. Emerging mechanisms of T-tubule remodelling in heart failure. Cardiovasc. Res. 2013;98:204–215. doi: 10.1093/cvr/cvt020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stølen T.O., Høydal M.A., Wisløff U. Interval training normalizes cardiomyocyte function, diastolic Ca2+ control, and SR Ca2+ release synchronicity in a mouse model of diabetic cardiomyopathy. Circ. Res. 2009;105:527–536. doi: 10.1161/CIRCRESAHA.109.199810. [DOI] [PubMed] [Google Scholar]
- 13.Swift F., Franzini-Armstrong C., Louch W.E. Extreme sarcoplasmic reticulum volume loss and compensatory T-tubule remodeling after Serca2 knockout. Proc. Natl. Acad. Sci. USA. 2012;109:3997–4001. doi: 10.1073/pnas.1120172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song L.S., Guatimosim S., Lederer W.J. Calcium biology of the transverse tubules in heart. Ann. N. Y. Acad. Sci. 2005;1047:99–111. doi: 10.1196/annals.1341.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrantini C., Coppini R., ter Keurs H.E. Impact of detubulation on force and kinetics of cardiac muscle contraction. J. Gen. Physiol. 2014;143:783–797. doi: 10.1085/jgp.201311125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Øyehaug L., Loose K.Ø., Louch W.E. Synchrony of cardiomyocyte Ca(2+) release is controlled by T-tubule organization, SR Ca(2+) content, and ryanodine receptor Ca(2+) sensitivity. Biophys. J. 2013;104:1685–1697. doi: 10.1016/j.bpj.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L., Louch W.E., Smith N.P. Calcium dynamics in the ventricular myocytes of SERCA2 knockout mice: a modeling study. Biophys. J. 2011;100:322–331. doi: 10.1016/j.bpj.2010.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louch W.E., Hake J., Sejersted O.M. Slow Ca2+ sparks de-synchronize Ca2+ release in failing cardiomyocytes: evidence for altered configuration of Ca2+ release units? J. Mol. Cell. Cardiol. 2013;58:41–52. doi: 10.1016/j.yjmcc.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Kekenes-Huskey P.M., Cheng Y., Michailova A.P. Modeling effects of L-type ca(2+) current and na(+)-ca(2+) exchanger on ca(2+) trigger flux in rabbit myocytes with realistic T-tubule geometries. Front. Physiol. 2012;3:351. doi: 10.3389/fphys.2012.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savio-Galimberti E., Frank J., Sachse F.B. Novel features of the rabbit transverse tubular system revealed by quantitative analysis of three-dimensional reconstructions from confocal images. Biophys. J. 2008;95:2053–2062. doi: 10.1529/biophysj.108.130617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres N.S., Sachse F.B., Bridge J.H. A modified local control model for Ca2+ transients in cardiomyocytes: junctional flux is accompanied by release from adjacent non-junctional RyRs. J. Mol. Cell. Cardiol. 2014;68:1–11. doi: 10.1016/j.yjmcc.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Lichter J.G., Sachse F.B. Cardiac resynchronization therapy reduces subcellular heterogeneity of ryanodine receptors, T-tubules, and Ca2+ sparks produced by dyssynchronous heart failure. Circ. Heart Fail. 2015;8:1105–1114. doi: 10.1161/CIRCHEARTFAILURE.115.002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sachse F.B., Torres N.S., Bridge J.H. Subcellular structures and function of myocytes impaired during heart failure are restored by cardiac resynchronization therapy. Circ. Res. 2012;110:588–597. doi: 10.1161/CIRCRESAHA.111.257428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crossman D.J., Ruygrok P.N., Cannell M.B. Changes in the organization of excitation-contraction coupling structures in failing human heart. PLoS One. 2011;6:e17901. doi: 10.1371/journal.pone.0017901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyon A.R., MacLeod K.T., Gorelik J. Loss of T-tubules and other changes to surface topography in ventricular myocytes from failing human and rat heart. Proc. Natl. Acad. Sci. USA. 2009;106:6854–6859. doi: 10.1073/pnas.0809777106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ang K.L., Shenje L.T., Galiñanes M. Limitations of conventional approaches to identify myocyte nuclei in histologic sections of the heart. Am. J. Physiol. Cell Physiol. 2010;298:C1603–C1609. doi: 10.1152/ajpcell.00435.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasqualin C., Gannier F., Maupoil V. Automatic quantitative analysis of t-tubule organization in cardiac myocytes using ImageJ. Am. J. Physiol. Cell Physiol. 2015;308:C237–C245. doi: 10.1152/ajpcell.00259.2014. [DOI] [PubMed] [Google Scholar]
- 28.Guo A., Song L.S. AutoTT: automated detection and analysis of T-tubule architecture in cardiomyocytes. Biophys. J. 2014;106:2729–2736. doi: 10.1016/j.bpj.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei S., Guo A., Song L.S. T-tubule remodeling during transition from hypertrophy to heart failure. Circ. Res. 2010;107:520–531. doi: 10.1161/CIRCRESAHA.109.212324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arora R., Aistrup G.L., Wasserstrom J.A. Regional distribution of T-tubule density in left and right atria in dogs. Heart Rhythm. 2017;14:273–281. doi: 10.1016/j.hrthm.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pizer S.M., Amburn E.P., Zimmerman J.B. Adaptive histogram equalization and its variations. Comput. Vis. Graph. Image Process. 1987;39:355–368. [Google Scholar]
- 32.Van Trees H.L. John Wiley & Sons, Inc.; New York: 2001. Detection, Estimation, and Modulation Theory, Part I; p. 716. [Google Scholar]
- 33.McNary T.G., Spitzer K.W., Sachse F.B. Mechanical modulation of the transverse tubular system of ventricular cardiomyocytes. Prog. Biophy. Mol. Biol. 2012;110:218–225. doi: 10.1016/j.pbiomolbio.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melo T.G., Almeida D.S., Pereira M.C. Disarray of sarcomeric alpha-actinin in cardiomyocytes infected by Trypanosoma cruzi. Parasitology. 2006;133:171–178. doi: 10.1017/S0031182006000011. [DOI] [PubMed] [Google Scholar]
- 35.Lichter J.G., Carruth E., Sachse F.B. Remodeling of the sarcomeric cytoskeleton in cardiac ventricular myocytes during heart failure and after cardiac resynchronization therapy. J. Mol. Cell. Cardiol. 2014;72:186–195. doi: 10.1016/j.yjmcc.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feldman A.M., Weinberg E.O., Lorell B.H. Selective changes in cardiac gene expression during compensated hypertrophy and the transition to cardiac decompensation in rats with chronic aortic banding. Circ. Res. 1993;73:184–192. doi: 10.1161/01.res.73.1.184. [DOI] [PubMed] [Google Scholar]
- 37.Jouannot P., Hatt P.Y. Rat myocardial mechanics during pressure-induced hypertrophy development and reversal. Am. J. Physiol. 1975;229:355–364. doi: 10.1152/ajplegacy.1975.229.2.355. [DOI] [PubMed] [Google Scholar]
- 38.Lyon A.R., MacLeod K.T., Gorelik J. Loss of T-tubules and other changes to surface topography in ventricular myocytes from failing human and rat heart. Proc. Natl. Acad. Sci. USA. 2009;106:6854–6859. doi: 10.1073/pnas.0809777106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frisk M., Ruud M., Louch W.E. Elevated ventricular wall stress disrupts cardiomyocyte t-tubule structure and calcium homeostasis. Cardiovasc. Res. 2016;112:443–451. doi: 10.1093/cvr/cvw111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen B., Li Y., Song L.S. β-Adrenergic receptor antagonists ameliorate myocyte T-tubule remodeling following myocardial infarction. FASEB J. 2012;26:2531–2537. doi: 10.1096/fj.11-199505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mørk H.K., Sjaastad I., Louch W.E. Slowing of cardiomyocyte Ca2+ release and contraction during heart failure progression in postinfarction mice. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H1069–H1079. doi: 10.1152/ajpheart.01009.2008. [DOI] [PubMed] [Google Scholar]
- 42.Lenaerts I., Bito V., Willems R. Ultrastructural and functional remodeling of the coupling between Ca2+ influx and sarcoplasmic reticulum Ca2+ release in right atrial myocytes from experimental persistent atrial fibrillation. Circ. Res. 2009;105:876–885. doi: 10.1161/CIRCRESAHA.109.206276. [DOI] [PubMed] [Google Scholar]
- 43.Sachse F.B., Savio-Galimberti E., Bridge J.H. Sub-micrometer anatomical models of the sarcolemma of cardiac myocytes based on confocal imaging. In: Altman R.B., Keith A., Hunter L., Murray T., Klein T.E., editors. Biocomputing 2008: Proceedings of the Pacific Symposium. 2008. pp. 390–401. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.