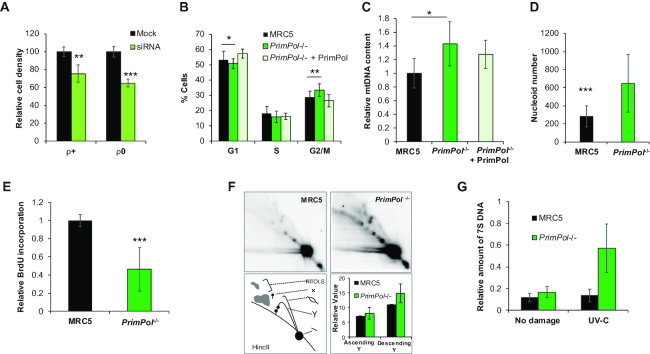
Figure 1.
PrimPol knockout cells proliferate normally, have increased mtDNA content but exhibit decreased replication rates. (A) Human Osteosarcoma cells (143B) treated with PrimPol siRNA or scrambled control were monitored using an IncuCyte™ microscope for 80 h with cells imaged every 3 h and cell density calculated (see Supplementary Figure S1). This shows the mean cell density at 60 h after RNAi from n = 3 experiments with standard deviation shown with error bars. (B) Cell cycle populations were measured in asynchronously growing MRC5 and PrimPol−/− cells or PrimPol−/− cells complemented with GFP-tagged PrimPol after PI staining and flow cytometry. (C) mtDNA copy number was analysed by Taqman probe qPCR in human MRC5 cells with and without PrimPol. (D) The mitochondrial network and mtDNA nucleoids were analysed by co-staining cells with Mitotracker (red), anti-DNA (green) and DAPI (blue) and ImageJ was used to analyse the number of mtDNA nucleoids per cell. (E) mtDNA replication was measured using BrdU incorporation, nuclear DNA replication was blocked using aphidicolin to allow mtDNA-specific analysis and cells were allowed to accumulate BrdU for 24 h. DNA was extracted, dot blotted and BrdU content quantified by anti-BrdU staining, this was then compared to total DNA content by the use of a mtDNA-specific radio-labelled probe, n = 3 independent repeats performed in triplicate. (F) mtDNA replication intermediates were viewed directly by 2D-AGE, total DNA was digested by HinCII and separated in two dimensions. DNA was Southern blotted and probed with an OH specific radio-labelled probe. Images were quantified using AIDA. (G) 7S DNA copy number was measured in undamaged and UV-C treated cells using southern blotting of whole cell DNA after PvuII digestion and probed with a 7S DNA and nuclear DNA specific radio-labelled probe, and quantified using AIDA software. Charts represent the mean of n≥3 independent experiments with error bars showing standard deviation, significance was measured using a Student's t-test, *P≤ 0.05, **P≤ 0.01, ***P≤ 0.001.