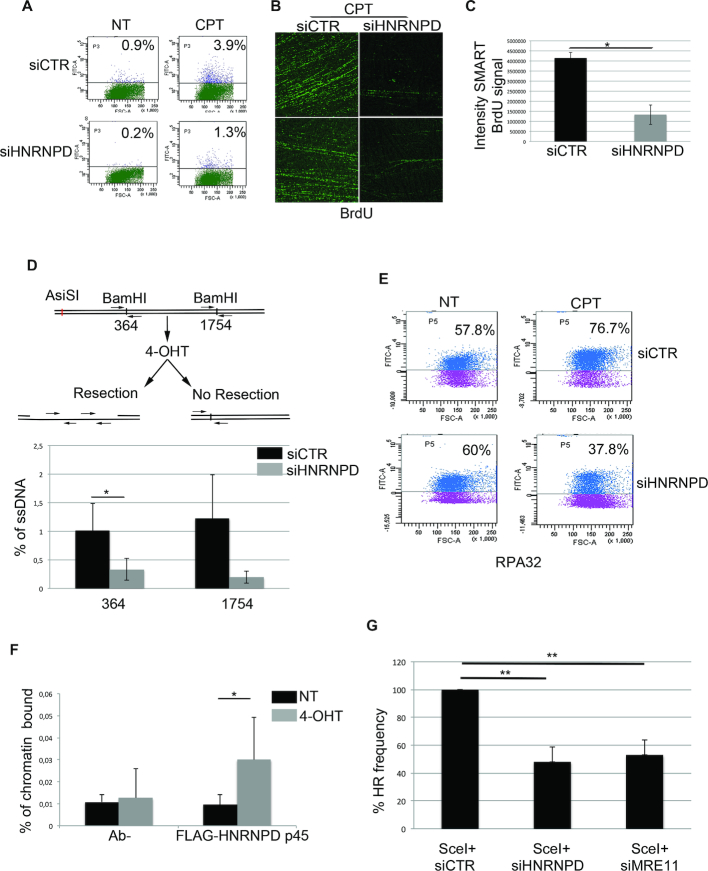
Figure 3.
HNRNPD regulates the DNA end-resection process. (A) FACS analysis of ssDNA formation in HeLa cells transfected with siCTR or siHNRNPD for 24 h, then pulse-labelled with 10 μM BrdU for additional 24 h and treated with 1 μM CPT for 2 h. HeLa cells were prepared for cytofluorimetric analysis in non-denaturing conditions in order to quantify the percentage of positive ssDNA cells. The untreated condition (no DNA damage) reveals the background level given in each cell type by the FITC-antibody detecting BrdU (in this case BrdU is entrapped within the double helix and cannot be recognized by the antibody). Such background values, subtracted to the values obtained following CPT treatment, indicate a ∼3-fold decrease in single strand DNA upon HNRNPD silencing. (B) To perform the SMART technique, HeLa cells (both siCTR and siHNRNPD) were pulse-labelled with 10 μM BrdU for 24 h then treated with 1 μM CPT for 2 h before spreading onto Silane Prep Slides. Immunofluorescence was performed with the BrdU antibody. Two representative images for each cell type (siCTR and siHNRNPD) are shown. (C) BrdU signal intensity of SMART technique analyzed by ImageJ software. Data represent the mean ± standard deviation (s.d.). (n = 2 independent experiments). *P-value <0.05. (D) HeLa ER-AsiSI were transfected with siCTR or siHNRNPD for 48 h followed by treatment with 300 nM of 4-OHT. The Real-time qPCR was performed on the genomic DNA with the indicated primers amplifying regions including the positions at 364 and 1754 bp downstream from the DSB, as depicted. The values of %ssDNA are reported as the mean ± s.d. (n = 4 independent experiments) and were calculated as follows: %ssDNA = 1/[(2ΔCt-1) + 0.5] × 100 (83). **P-value <0.01; *P-value <0.05. (E) HeLa cell lines were transfected with the indicated siRNAs for 48 h treated or not with 1 μM CPT for 2 h; before fixative, cells were pre-extracted with the CSK + 0.1% Triton X100 + 0.3 μg/ml RNase A followed by incubation with indicated antibody. RPA32 values were analyzed through flow cytometry. (F) ChIP experiments of HeLa ER-AsiSI, treated or mock treated with 300 nM 4-OHT for 1 h, were performed by using either an HNRNPD antibody or control IgGs (Ab-). The HNRNPD chromatin binding ability was measured, as a percentage of immunoprecipitated input, from qPCR values of an 80 bp amplicon including the AsiSI site. Data represent the mean ± s.d. (n = 4 independent experiments). *P-value <0.05. (G) HeLa DR-GFP cells were transfected with the plasmid encoding the SceI restriction enzyme in presence of siCTR, siHNRNPD or siMRE11 followed by FACS analysis measurement of GFP levels used to calculate %HR frequency compared with siCTR which was set as 100%. Data represent the mean ± s.d. (n = 3 independent experiments), **P-value <0.01.