Abstract
Transcription-coupled nucleotide excision repair (TC-NER) is a dedicated DNA repair pathway that removes transcription-blocking DNA lesions (TBLs). TC-NER is initiated by the recognition of lesion-stalled RNA Polymerase II by the joint action of the TC-NER factors Cockayne Syndrome protein A (CSA), Cockayne Syndrome protein B (CSB) and UV-Stimulated Scaffold Protein A (UVSSA). However, the exact recruitment mechanism of these factors toward TBLs remains elusive. Here, we study the recruitment mechanism of UVSSA using live-cell imaging and show that UVSSA accumulates at TBLs independent of CSA and CSB. Furthermore, using UVSSA deletion mutants, we could separate the CSA interaction function of UVSSA from its DNA damage recruitment activity, which is mediated by the UVSSA VHS and DUF2043 domains, respectively. Quantitative interaction proteomics showed that the Spt16 subunit of the histone chaperone FACT interacts with UVSSA, which is mediated by the DUF2043 domain. Spt16 is recruited to TBLs, independently of UVSSA, to stimulate UVSSA recruitment and TC-NER-mediated repair. Spt16 specifically affects UVSSA, as Spt16 depletion did not affect CSB recruitment, highlighting that different chromatin-modulating factors regulate different reaction steps of the highly orchestrated TC-NER pathway.
INTRODUCTION
Eukaryotic gene transcription by RNA Polymerase II (Pol II) is crucial for proper cell function. However, different types of DNA lesions can damage the Pol II template, thereby severely impeding or even stalling the progression of elongating Pol II. These transcription-blocking DNA lesions (TBLs) can originate from endogenous or exogenous sources; for example, metabolic byproducts may induce oxidative DNA damage or ultraviolet (UV)-light-induced helix-distorting lesions such as cyclobutane pyrimidine dimers (CPDs) (1–3). TBLs pose a direct problem for cellular homeostasis due to a lack of newly synthesized RNA or to the formation of mutant RNA molecules. In addition, prolonged stalling of Pol II may result in collisions with advancing replication forks and may induce R-loop formation (4). TBLs can therefore cause genome instability, severe cellular dysfunction, premature cell death and senescence, which finally may result in DNA damage induced, accelerated aging (5–7).
To overcome these cytotoxic TBLs, cells are endowed with transcription-coupled nucleotide excision repair (TC-NER). TC-NER is a dedicated branch of the nucleotide excision repair pathway that specifically repairs TBLs in the transcribed strand of active genes, thereby resolving lesions that stall RNA Pol II and subsequently allowing transcription to restart (4,8). The importance of TC-NER is best shown by its causative link with the Cockayne Syndrome (CS) and the UV-sensitivity syndrome (UVSS) (6,9,10). CS is caused by mutations in Cockayne Syndrome protein A (CSA) and Cockayne Syndrome protein B (CSB) (11,12), while mutations in UVSSA give rise to UVSS (13–15). Despite a similar deficiency in the repair of UV-induced TBLs, the CS and UVSS phenotypes are strikingly different (6,9,10). CS is characterized by photosensitivity, growth failure, progressive neurodevelopmental defects and premature aging (10,16), while UVSS has a far less severe phenotype, which is restricted to cutaneous photosensitivity, such as freckling and pigmentation abnormalities (9).
The recognition of lesion-stalled Pol II by CSB is assumed to be the initiating signal for TC-NER (17–19). In unperturbed conditions, the transcription elongation factor CSB transiently interacts with elongating Pol II; however, this interaction becomes more stable when Pol II is stalled at a TBL (18,20). In line with this, recent cryo-EM studies of Rad26, the yeast homolog of CSB, show that it binds DNA upstream of Pol II, where it has a key role in lesion recognition (19). Through its adenosine triphosphatase activity, Rad26 facilitates forward translocation of Pol II over naturally occurring pause sites or less bulky lesions. However, Rad26 cannot translocate Pol II over bulky TBLs (19). This prolonged binding of CSB to lesion-stalled Pol II is thought to be one of the first steps in the assembly of the TC-NER complex, as for example shown by the CSB-dependent CSA translocation to the nuclear matrix following UV-induced DNA damage (21). CSA forms an E3-ubiquitin ligase complex with DDB1, Cul4A, ROC1/Rbx1 (22,23), and is involved in the ubiquitylation and subsequent degradation of CSB upon UV irradiation (24). The UV-induced degradation of CSB is counteracted by the deubiquitylating enzyme USP7, which is recruited by the TC-NER factor UV-Stimulated Scaffold Protein A (UVSSA) (13,14). Furthermore, UVSSA plays a role in the restoration of the hypo-phosphorylated form of Pol II (Pol IIa) (13) and in UV-induced ubiquitin modifications of Pol II (15), but both effects might be indirect. Recently, it was suggested that UVSSA also plays an important role in the recruitment of the transcription factor II H (TFIIH) via a direct interaction with P62 (15,25). TFIIH subsequently unwinds a stretch of ∼30 nt surrounding the damage site and is, in combination with XPA and RPA, responsible for damage verification and the orientation of the XPF/ERCC1 and XPG endonucleases, thereby playing an important role in the DNA strand specificity. Following excision of the damaged DNA, the resulting single-stranded gap is filled by DNA synthesis and sealed by DNA ligases (6).
Despite significant advances, the regulation and recruitment mechanisms of TC-NER factors to lesion-stalled Pol II are thus far not fully understood and such understanding is required for proper comprehension of the TC-NER mechanism and its disease etiology. For example, the exact recruitment mechanism of UVSSA remains under debate. Like CSB, UVSSA has affinity for Pol II in unperturbed conditions (14,18,26), and it has been suggested that this interaction is stabilized following DNA damage (13). Although UVSSA interacts with CSA (27), UVSSA accumulation at sites of UV-induced DNA damage is a CSA- and CSB-independent process (14). In contrast, the UV-induced UVSSA translocation to chromatin observed in cell fractionation assays was shown to depend on CSA (27).
To increase our understanding of the spatiotemporal build-up of the TC-NER complex and its molecular mechanism, we compared the accumulation kinetics of different TC-NER factors in living cells and studied the UVSSA recruitment in TC-NER-deficient cells in a quantitative manner. Our analysis showed that UVSSA recruitment to DNA damage occurs in a CSA- and CSB-independent manner. In addition, UVSSA deletion mutants showed that UVSSA binding to CSA and recruitment to TBLs are mediated by distinct domains: the Vps/Hrs/STAM (VHS) domain and the domain of unknown function 2043 (DUF2043), respectively. Using these separation-of-function mutants of UVSSA, in combination with quantitative interaction proteomics, we identified the Spt16 subunit of the H2A/H2B chaperone FACT (facilitates chromatin transcription) to be involved in the UVSSA recruitment. Spt16 is recruited early in the TC-NER reaction in a UVSSA-independent manner, thereby stimulating excision of the TBLs and subsequent transcription restart after DNA damage removal. Our work establishes Spt16 as an important regulator of TC-NER-mediated repair and provides new insights into the different mechanisms involved in the recognition of lesion-stalled Pol II and how the remodeling of chromatin fine-tunes the regulation of the different stages of TC-NER.
MATERIALS AND METHODS
Plasmid constructs
GFP-tagged UVSSA deletion mutants of the DUF2043 and nuclear localization signal (NLS) domains amino acids 495–709 (ΔDUF), DUF2043 domain amino acids 495–605 (ΔDUFonly), C-terminal NLS amino acids 645–709 (ΔNLS) and VHS domain amino acid 1–152 (ΔVHS) domain were made by polymerase chain reaction (PCR) amplification on pLenti CMV Hygro vector (28), containing either a full length C1-UVSSA construct or an N2-UVSSA (for ΔVHS) construct, with Phusion High-Fidelity DNA polymerase (M0530, New England Biosciences) using the following primers: ΔDUF Forward 5′-CACCATGGTGAGCAAGGGCGAG-3′, ΔDUF Reverse 5′-CTATGCTGCCAGCTTCTGGGCCTC-3′, ΔVHS Forward 5′-CACCATGTTTCAAGACACGAATGCTCGGAGT-3′, ΔVHS Reverse 5′-TTACTTGTACAGCTCGTCCAT-3′, ΔNLS Forward 5′-CACCATGGTGAGCAAGGGCGAG-3′ and ΔNLS Reverse 5′-GCTGTACCTGGATGAGCCGAGAT-3′. PCR products were gel purified, to prevent contamination of later PCR reactions with template DNA, and subsequently subcloned into pENTR™/D-TOPO® vector using the pENTR™ directional TOPO® Cloning kit (Invitrogen). To generate the ΔDUFonly mutant the following primers were used to amplify the complete GFP-UVSSA construct in pENTR4-GFP-C1 (w392–1) lacking the DUF2043 domain: ΔDUFonly Forward 5′-phos-AGGGCTCGTGAGCAGCGGCG-3′ ΔDUFonly Reverse 5′-phos-TGCTGCCAGCTTCTGGGCCTCC-3′. The obtained PCR fragment was used in a subsequent T4 ligation reaction to reassemble the ΔDUFonly mutant in pENTR4-GFP-C1. All constructs were cloned by recombination into the pLenti CMV Hygro destination vector (Addgene, plasmid ID: #17454) using the Gateway LR Clonase II Enzyme Mix (Invitrogen).
Cell line generation
Full length GFP-UVSSA (14) or UVSSA deletion mutants (GFP-UVSSA ΔDUF, ΔDUFonly, ΔNLS and UVSSA ΔVHS-GFP) expressing cell lines were generated by lentiviral transduction of the indicated constructs. To that end, third-generation lentiviruses were made in HEK293T cells and were used to transduce UVSS-A (TA24) SV40-immortalized cells. Fibroblasts originating from NER patients (SV40 transformed) were complemented with the respective deficient NER protein as described: GFP-CSB in CS-B (CS1AN) (18), CSA-Flag-GFP in CS-A (CS3BE) (29), XPC-GFP in XP-C (XP4PA) (30), GFP-XPA in XP-A (XP20S) (31), GFP-XPB in XP-B (XPCS2BA) (32). Vh10 (hTert) cells stably expressing GFP-DDB2 were described before (33). The generation of U2OS cells stably expressing GFP-tagged Spt16 or SSRP1 was described before (34), UVSS-A (TA24) cells expressing GFP-tagged Spt16 were generated in a similar approach. TA24 GFP-Spt16 cells were complemented with FLAG-tagged UVSSA by lentiviral transduction. Gateway LR Clonase (Invitrogen) was used to recombine UVSSA-Flag from pENTR4 no ccDB (686–1, Addgene, plasmid ID: #17424) (14) to pLenti CMV Puro Dest (w118–1, Addgene, plasmid ID: #17452). The generated, rescued cell line was subjected to a combination of selection by Puromycin (2.5 μg/ml) for UVSSA-Flag and Hygromycin (5 μg/ml) for Spt16-GFP. GFP-H2A (34) was stably expressed in HeLa cells (34) or in UVSS-A (TA24) cells by transfection using X-treme Gene HP (Roche) according to the manufactures protocol. Cells stably expressing GFP-H2A were selected using 0.5 mg/ml G418 and FACS sorting.
Cell culture
TA24 (UVSS-A), CS1AN (CS-B), CS3BE (CS-A), XP4PA (XP-C), XP20S (XP-A), XPCS2BA (XP-B), HeLa, Vh10 and U2OS cell lines were cultured in a 1:1 ratio of Dulbecco's modified Eagle's medium (DMEM) and Ham's F10 (Invitrogen) containing 10% fetal calf serum (FCS, Biowest) and antibiotics at 37°C and 5% CO2. For SILAC labeling, cells were cultured for 2 weeks in DMEM without lysine, arginine or leucine (AthenaES) supplemented with antibiotics, 10% dialyzed FCS (Invitrogen) and 105 μg/ml leucine (Sigma) and either 73 μg/ml light [12C6]-lysine and 42 μg/ml [12C6,14N4]-arginine (Sigma) or with heavy [13C6]-lysine and [13C6,15N4]-arginine (Cambridge Isotope Laboratories) at 37°C and 5% CO2.
RNA interference
Transient siRNA-mediated knock-down was achieved using Lipofectamine RNAiMAX (Invitrogen) transfection, according to the manufacturer’s instruction. The siRNA oligonucleotides used, (Thermo Fisher Scientific) were as follows: CTRL (D-001210–05-20) 5′-UGGUUUACAUGUCGACUAA-3′, Spt16 (L-009517–00) 5′-AGUCUAAUGUGUCCUAUAA-3′, 5′-GCAUAUACCAUCGCUGUAA-3′, 5′-ACACGGAUGUGCAGUUCUA-3′, 5′-GUACAGCAAUUGGCGGAAA-3′, SSRP1 (L-011783–00), 5′-GCUCUGGGCCAUGGACUUA-3′, 5′-GGAGUUCAACGACGUCUAU-3′, 5′-CGAUGAAUAUGCUGACUCU-3′, 5′-AAGAAGAACUAGCCAGUAC-3′, UVSSA (J-0243197–23-0002) 5′-GCUCGUGGAUCCAGCGCUU-3′, Nap1 L1 (L-017274–01-0005), 5′-UAACCAUAGUUCAUCGAAAUU-3′, 5′-GCGUAUAAUCCCAAGAUCAUU-3′, 5′-GUUAAGGCAUAUUGAGUUAUU-3′, 5′-GGAACGAGAUGCUAUACU-3′
Clonogenic survival assay
Cells were seeded in triplicate in 6-well plates (300 cells/well) and treated with a single dose of the indicated UV-C dose (254 nm; Philips TUV lamp) 1 day after seeding. After 1 week, colonies were fixed and stained in 50% methanol, 7% acetic acid and 0.1% Coomassie blue and subsequently counted with the Gelcount (Oxford Optronix, Software Version 1.1.2.0). The survival was plotted as the mean percentage of colonies detected following the indicated UV-C dose compared to the mean number of colonies from the non-irradiated samples.
Live-cell confocal laser-scanning microscopy
Confocal laser-scanning microscopy images were obtained with a Leica SP5 confocal microscope using a 100x quartz objective for local UV-damage induction. Local DNA damage infliction for kinetic studies of GFP-tagged protein accumulation was performed using a 266 nm UV-C (2 mW pulsed (7.8 kHz) diode pumped solid-state laser (Rapp OptoElectronic, Hamburg) as described previously (14,35). Briefly, cells were grown on quartz cover slips and were imaged and irradiated through a 100 × 1.2 numerical aperture (NA) Ultrafluar quartz objective. During microscopy, cells were kept at 37°C and 5% CO2. Images were acquired using the LAS AF software (Leica) and the fluorescence intensity at the damage area was recorded over time, background corrected and normalized to pre-damage fluorescence levels to quantify accumulation kinetics. H2A exchange on UV-C induced DNA damage was performed as described previously (34). In short, half of the nucleus was photobleached by a 488 nm laser and local UV-C damage was subsequently induced in the bleached area. The recovery of fluorescence, representing histone exchange, on the UV-C damaged area and non-damaged area was quantified. Fluorescence intensities were background corrected and the fluorescence on the UV-C damaged area was normalized to the fluorescence for the non-damaged area. The indicated number of cells originate from at least two experiments and the results were pooled and plotted as the mean fluorescence intensity ± SEM.
Immunofluorescence
Cells were grown on 24-mm coverslips and fixed using 2% paraformaldehyde supplemented with triton X-100. Subsequently cells were permeabilized with phosphate buffered saline (PBS) containing 0.1% triton X-100. Coverslips were washed with PBS containing 0.15% glycine and 0.5% bovine serum albumin (BSA) and incubated with primary antibody, FLAG M2 (1:1000) for 1–2 h at room temperature. Cells were washed three times and two times for 10 min with 0.1% triton X-100 and once with PBS containing 0.15% glycine and 0.5% BSA. To visualize primary antibodies, coverslips were incubated for 1 h with secondary antibodies labeled with ALEXA fluorochrome 594 (Invitrogen). Again cells were washed with 0.1% Triton X-100 and PBS+. Subsequently coverslips were embedded in 4′,6-diamidino-2-phenylindole (DAPI) containing Vectashield mounting medium (Vector Laboratories). Images were obtained using a Zeiss LSM700 microscope equipped with a 63 × oil Plan-apochromat 1.4 NA oil immersion lens (Carl Zeiss Microimaging Inc.).
TC-NER specific unscheduled DNA synthesis
The amplified unscheduled DNA synthesis (UDS) assay was performed as described (36). Briefly, XP186LV, XPC-deficient cells, seeded on 24-mm coverslips 4 days prior to the experiment were transfected with siRNA 2 days later. One day following transfection the medium was replaced by low-serum containing medium (Ham’s F10 supplemented with 0.5% FCS) to reduce the number of cells in S-phase. For global UV-C irradiation (8 J/m2), a 254 nm germicidal lamp (Philips) was used. Following irradiation, cells were labeled with medium (Ham’s F10 supplemented with 0.5% dialyzed FCS) containing 5-ethynyl,2′-deoxyuridine (EdU, 20 μM, ThermoFisher Scientific) and 2′-Deoxy-5-fluorouridine (Floxuridine, 1 μM, Sigma-Aldrich) for 7 h. Subsequently, cells were fixed in 3.6% formaldehyde (Sigma-Aldrich) in PBS with 0.5% Triton X-100 (Sigma-Aldrich). EdU incorporation was visualized using a combination of the Click-iT reaction (Invitrogen) and Tyramide Signal Amplification (ThermoFisher Scientific). The Click-it reaction was performed as described in the manufactures protocol using Azide-PEG3-Biotin Conjugate (20 μM in Dimethylsulfoxide (DMSO), Jena Bioscience), 1× Click-it reaction buffer (ThermoFisher Scientific), Copper(III)sulphate (0.1 M) and 10× Reaction buffer additive (ThermoFisher Scientific). The tyramide-based amplification was conducted as described in the manufactures protocol by using the HRP-Streptavidin conjugate (500 μg/ml, ThermoFisher Scientific) and Alexa-Fluor 488 nm labeled tyramide (ThermoFisher Scientific). Cover slips were embedded in DAPI containing Vectashield mounting medium (VectorLaboratories) and sealed using nail polish, and visualized using a Zeiss LSM700 microscope equipped with a 40× oil Plan-Apochromat 40 0.6–1.3 NA oil immersion lens (Carl Zeiss Micro imaging Inc.). TC-NER-UDS signal was quantified by analyzing at least six fields for each condition Mean nuclear fluorescence signals were quantified using ImageJ software (Version 1.48) (37). Sample analysis was performed as described (36). The mean nuclear fluorescence signal ± standard error of the mean is shown.
In vivo cross-linking and immunoprecipitation
The cross-linked immunoprecipitation procedure has been described previously (14,34). Briefly, in vivo cross-linking was performed by adding 1% formaldehyde to the culture medium for 10 min at 4°C. Cross-linked cells were scrapped and chromatin was purified. Finally, the nuclear suspension was sonicated using the Bioruptor Sonicator (Diagenode) with 6 cycles of 30 s on and 60 s off. For immunoprecipitations, equal amounts of cross-linked chromatin from all samples were incubated in Radioimmunoprecipitation assay buffer (RIPA) with 30 μl GFP-Trap-A agarose bead slurry (ChromoTek), overnight at 4°C. Beads were collected by centrifugation, washed five times with RIPA buffer and GFP-tagged proteins were de-crosslinked and eluted by incubation at 95°C for 30 min in 2× Laemmli sodium dodecylsulphate (SDS) sample buffer. Samples were analyzed by western blot and loaded to 4–15% Mini-PROTEAN TGX™ Precast Protein Gels (BioRad). Gels were fixed and stained by Roti-blue (Carl Roth GmbH) according to the manufacturer’s protocol.
For native IP with Benzonase, cells were lysed in IP buffer (30 mM hydroxyethyl-piperazineethane-sulfonic acid (HEPES) pH 7.5, 130 mM NaCl, 1 mM MgCl2, 0.5% Triton X-100 and protease inhibitor cocktail) for 10 min at 4 °C. Subsequently, cells were sonicated using the Bioruptor Sonicator (Diagenode) with 10 cycles of 15 s on and 45 s off, and 500 U of Benzonase (Sigma) was added to the lysates. Following 1 h of incubation, the lysates were cleared by centrifugation and the supernatants were subjected to immunoprecipation with GFP-Trap-A agarose beads overnight at 4°C. Beads were collected by centrifugation, washed five times with IP buffer and GFP-tagged proteins were eluted by incubation at 95°C for 5 min in 2× Laemmli SDS sample buffer.
Western blot and antibodies
Lysates were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to a Polyvinylidene difluoride (PVDF) membrane (0.45 μm). Membranes were blocked with 5% milk in PBS for 1 h at room temperature and incubated with primary antibodies raised against GFP (Roche, 11814460001), CSA/ERCC8 (Abcam, ab137033), USP7 (Bethyl, A300–033A), Spt16, SSRP1 (Santa Cruz Biotechnology, sc-28734 and sc-74536, respectively), Spt16 (Abcam, Ab56855) or Tubulin (Sigma Aldrich, clone B-5–1-2). Secondary antibodies coupled to IRDyes (LI-COR) were used to visualize proteins using an Odyssey CLx infrared scanner (LiCor).
Mass spectrometry
SDS-PAGE gel lanes were cut into 2-mm slices and subjected to in-gel reduction with dithiothreitol, alkylation with iodoacetamide and digestion with trypsin (sequencing grade; Promega), as described previously (14). Nanoflow liquid chromatography tandem mass spectrometry (nLC-MS/MS) was performed on an EASY-nLC coupled to a Q Exactive mass spectrometer (Thermo) or to an Orbitrap Fusion Tribid mass spectrometer (Thermo), both operating in positive mode. Peptide mixtures were trapped on a ReproSil C18 reversed phase column (Dr Maisch; 1.5 cm × 100 μm) at a rate of 8 μl/min. Peptides were separated on a ReproSil-C18 reversed-phase column (Dr Maisch; 15 cm × 50 μm) using a linear gradient of 0–80% acetonitrile (in 0.1% formic acid) for 170 min at a rate of 200 nl/min. The elution was directly sprayed into the electrospray ionization source of the mass spectrometer. Spectra were acquired in continuum mode; fragmentation of the peptides was performed in data-dependent mode by HCD (Q Exactive) or CID (Orbitrap Fusion).
Raw mass spectrometry data were analyzed using the MaxQuant software suite (version 1.4.1.2) (38). A false discovery rate of 0.01 for proteins and peptides and a minimum peptide length of seven amino acids were set. The Andromeda search engine was used to search the MS/MS spectra against the Uniprot database (taxonomy: Homo sapiens, release June 2013). A maximum of two missed cleavages was allowed. The peptide tolerance was set to 10 ppm and the fragment ion tolerance was set to 20 mmu for HCD spectra (Q Exactive) or to 0.6 Da for CID spectra (Orbitrap Fusion). The enzyme specificity was set to ‘trypsin’, and cysteine carbamidomethylation was set as a fixed modification. SILAC protein ratios were calculated as the median of all peptide ratios assigned to the protein. In addition, a posterior error probability for each MS/MS spectrum below or equal to 0.1 was required. In case the identified peptides of two proteins were the same or the identified peptides of one protein included all peptides of another protein, these proteins were combined by MaxQuant and reported as one protein group. Before further statistical analysis, known contaminants and reverse hits were removed.
RESULTS
UVSSA accumulates independently of CSA and CSB on UV-C induced DNA damage
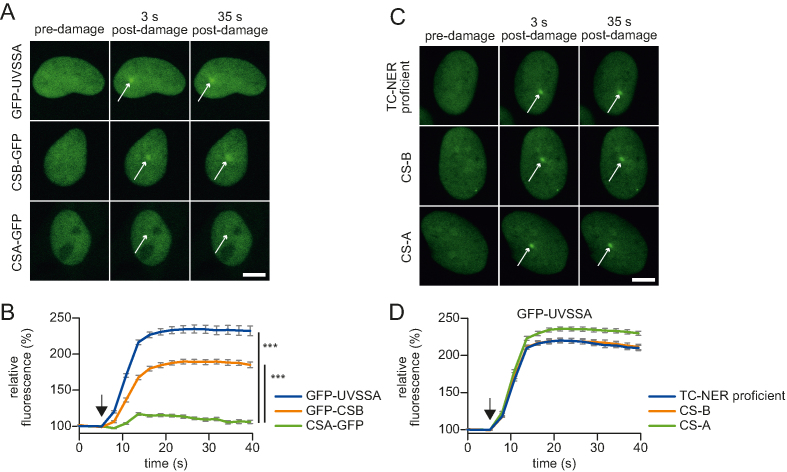
To acquire more insights into the recruitment mechanism of UVSSA, we first quantified its accumulation at sites of DNA damage and compared this to the recruitment kinetics of the other TC-NER initiating factors, CSA and CSB. For this purpose, we used TC-NER-deficient patient cell lines that are mutated in either CSA, CSB or UVSSA and are functionally complemented by the stable expression of GFP-tagged versions of the respective full-length TC-NER factors (14,18,29) (Supplementary Figure S1A and B). Accumulation kinetics of these TC-NER factors at sites of local UV-C laser-induced (266 nm) DNA damage were determined using quantitative live-cell confocal imaging (35). GFP-UVSSA and GFP-CSB recruitment at sites of locally UV-induced DNA damage (LUD) was clearly visible and showed a similar, swift, 2-fold accumulation (Figure 1A and B). Interestingly, despite the fact that CSA is a crucial TC-NER factor (5) and has been shown to directly interact with both UVSSA and CSB (13,24,27) (Figure 2C), its accumulation at sites of LUD was barely detectable (Figure 1A and B). This might be explained by a transient binding of CSA to the TC-NER complex. Furthermore, the almost complete absence of CSA at LUD makes it unlikely that UVSSA recruitment to the TC-NER complex is mediated via a direct protein–protein interaction with CSA. In line with this assumption, we have previously shown that UVSSA accumulation can still be detected at sites of UV-C-induced DNA damage in the absence of functional CSA and CSB (14). However, as these experiments do not rule out that CSA or CSB might have more subtle effects on UVSSA recruitment kinetics, for example, reduced accumulating rates or levels, we determined the accumulation of GFP-UVSSA in time in a quantitative manner in CSA- and CSB-deficient cells and compared this to that in TC-NER-proficient cells (complemented UVSS-A patient cell line) (Figure 1C and D). GFP-UVSSA was recruited with the same kinetics in TC-NER-proficient cells as in CSA- or CSB-deficient cells, indicating that UVSSA accumulation is not influenced by CSA or CSB activities.
Figure 1.
Accumulation kinetics of TC-NER factors reveal a CSA independent UVSSA recruitment. (A) Representative images of live cell imaging analysis of GFP-UVSSA, GFP-CSB or CSA-GFP at the indicated time points following local UV-C laser (266 nm) induced damage (LUD) in a sub-nuclear region (indicated by a white arrow); scale bar: 7.5 μm. (B) Relative accumulation of the indicated GFP-tagged TC-NER factors. GFP fluorescence intensity at LUD was quantified over time and normalized to pre-damage intensity set at 100 at t = 0 (n = 25 cells of two independent experiments, mean ± SEM). The moment of damage induction is indicated with a black arrow. A one-way Anova test was performed and P-values <0.001 (***) are depicted. (C) Representative images of GFP-UVSSA accumulation at the indicated time points at LUD (indicated by a white arrow). GFP-UVSSA is expressed to functionally complement UVSS-A cells (TC-NER proficient cells) or GFP-UVSSA expressed in CS-A and CS-B cell lines; scale bar: 7.5 μm. (D) Relative accumulation of GFP-UVSSA in the indicated cell lines. GFP fluorescence intensity at LUD of was quantified over time and normalized to pre-damage intensity set at 100 at t = 0 (n = 30 cells, two independent experiments, mean ± SEM). The moment of damage induction is indicated with a black arrow.
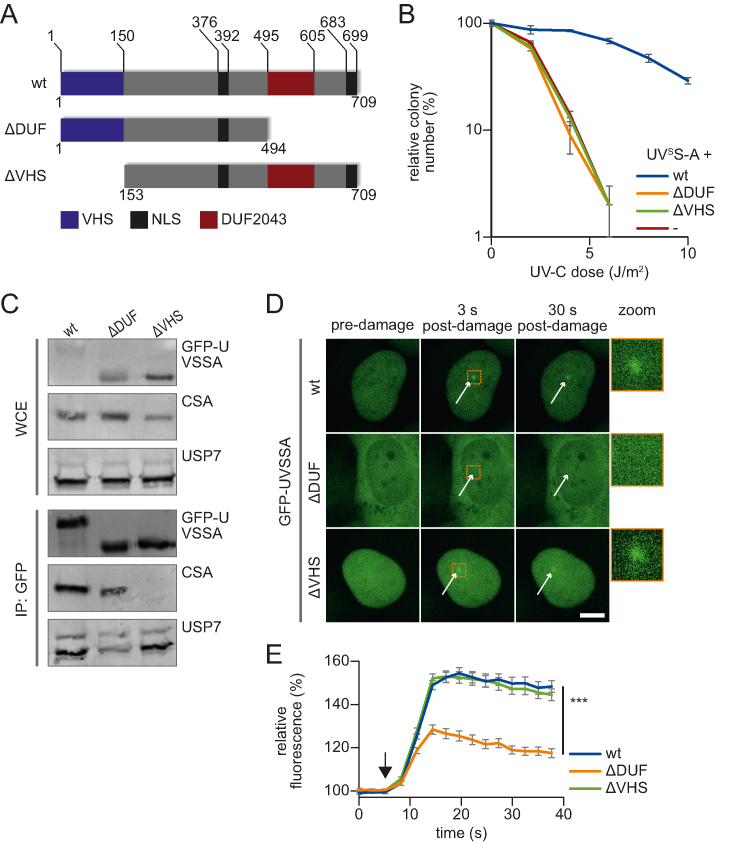
Figure 2.
CSA interaction and recruitment to DNA damage is mediated by distinct UVSSA domains. (A) Schematic overview of the protein domains present in UVSSA and the used UVSSA deletion mutants that either lack the VHS domain (ΔVHS) or the DUF2043 domain (ΔDUF). NLS: nuclear localization signal. (B) UV sensitivity of UVSS-A cells (-) and UVSS-A cells complemented with GFP-UVSSA (wt), GFP-UVSSA ΔDUF (ΔDUF) or UVSSA ΔVHS-GFP (ΔVHS) was determined by their colony-forming ability, following irradiation with the indicated UV-C doses. The percentage of surviving colonies is plotted against the UV-C dose. The number of colonies counted at 0 J/m2 is set as 100% survival. Data represents the experiment conducted in triplicate and error bars represent SEM. (C) Whole-cell extracts (WCE) of UVSS-A patient cells stably expressing the indicated constructs were subjected to GFP immunoprecipitation. Western blot analysis of the immunoprecipitated proteins was performed using GFP, CSA or USP7 antibodies. WCE: whole-cell extract, IP: Immunoprecipitate. (D) Representative images of live cell imaging analysis of GFP-UVSSA or ΔDUF and ΔVHS mutants following local UV-C laser (266 nm) induced damage (indicated by a white arrow); scale bar: 7.5 μm. Right panel: 4× zoomed image to visualize accumulation at 3 s post-damage induction. (E) GFP fluorescence intensity of the indicated constructs at LUD was quantified over time and normalized to pre-damage intensity set at 100 at t = 0 (n = 30 cells of two independent experiments, mean ± SEM). A one-way Anova test was performed and P-values < 0.001 (***) are depicted. The moment of damage induction is indicated with a black arrow.
The VHS and DUF2043 domains of UVSSA have distinct functions and are both required for TC-NER
To gain insight into the mechanism of UVSSA recruitment to TBLs, we tested which domain of UVSSA is involved in this process. Therefore, we stably expressed two GFP-tagged UVSSA deletion mutants in UVSSA-deficient TA24 cells, in which either the C-terminal DUF2043 domain (ΔDUF) or the N-terminal VHS domain (ΔVHS) (15) was deleted (Figure 2A). In contrast to expression of the full-length GFP-UVSSA (wt), cells expressing either ΔDUF or ΔVHS UVSSA mutants showed similar UV-hypersensitivity as UVSSA-deficient cells (Figure 2B), indicating that both the DUF2043 and VHS domains are essential for TC-NER. In line with previous data that mapped the CSA interaction domain to the N-terminus of UVSSA (27), immunoprecipitation experiments showed that the deletion of the N-terminal VHS domain resulted in the complete loss of CSA interaction, while this interaction remained unaffected in the ΔDUF mutant (Figure 2C and Supplementary Figure S2A). Of note, the ΔVHS mutant could still interact with the known UVSSA interaction partner USP7, indicating that the ΔVHS mutant is at least partially functional. Like the wt UVSSA, the ΔVHS mutant showed strict nuclear localization (Figure 2D). However, the ΔDUF mutant featured an additional cytoplasmic localization, which may have been caused by the deletion of the predicted C-terminal NLS (15). Despite its partial cytoplasmic localization, a significant fraction of the ΔDUF mutant remained present in the nucleus, in line with the retained interaction with the nuclear TC-NER factor CSA (21) (Figure 2C). Subsequently, we quantified the recruitment kinetics of these UVSSA mutants to DNA damage. While the ΔVHS mutant accumulated to the same level as wt UVSSA, the recruitment of the ΔDUF mutant to sites of local DNA damage was severely reduced (Figure 2D and E). To test whether the partial cytoplasmic localization of the ΔDUF mutant influences the UVSSA accumulation at DNA lesions, we generated two additional mutants in which either the DUF2043 domain alone (ΔDUFonly) or the C-terminal-predicted NLS domain (ΔNLS) was removed (Supplementary Figure S2B and C). In contrast to the ΔDUF mutant, the ΔDUFonly mutant was specially localized to the nucleus (Supplementary Figure S2D). However, deletion of the DUF2043 domain still severely reduced the accumulation of UVSSA at DNA damage (Supplementary Figure S2E). Deletion of the NLS alone (ΔNLS) did not affect the UVSSA accumulation. Surprisingly, this ΔNLS mutant was also localized mainly in the nucleus, suggesting that the region that is deleted in the ΔDUF mutant, located between the DUF2043 domain and the predicted NLS sequence, is important for the nuclear localization of UVSSA (Supplementary Figure S2B and D). Together our data show a clear separation-of-function of the UVSSA domains; the VHS domain is crucial for CSA interaction, while the DUF2043 domain plays an important role during the UVSSA recruitment to DNA damage. Importantly, in line with the unaffected UVSSA accumulation in CS-A cells, these data further show that UVSSA recruitment to TBLs is a CSA-independent process.
Identification of UVSSA-interacting proteins
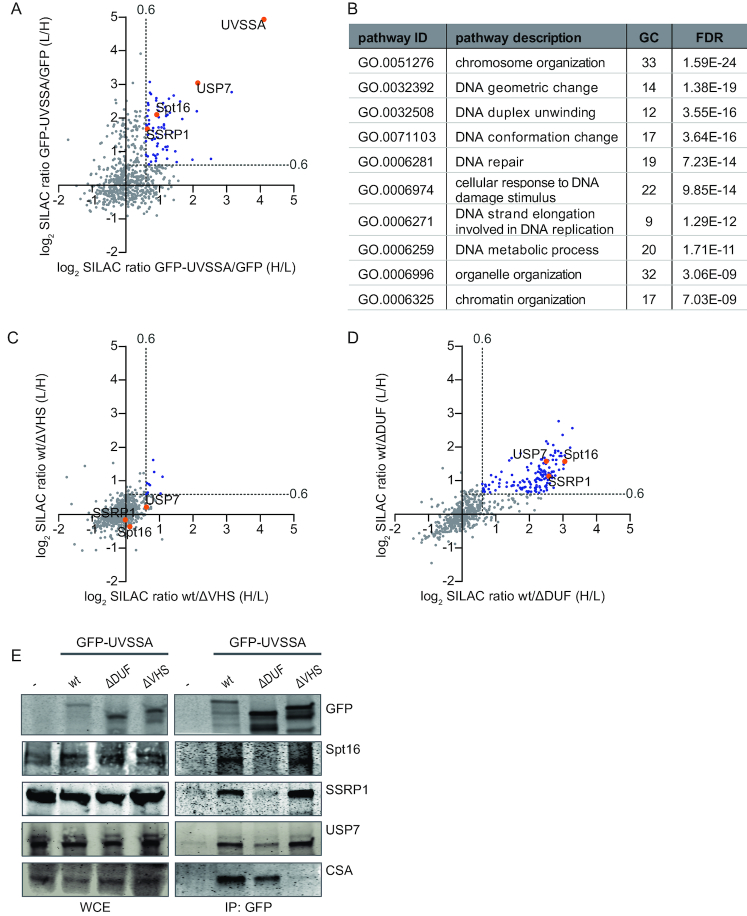
To identify proteins that are involved in recruiting UVSSA to DNA damage, we set out to identify UVSSA-interacting proteins using SILAC-based quantitative interaction proteomics. UVSSA-containing protein complexes were isolated using GFP-nanotrap pulldowns (39) in UVSS-A cells stably complemented with GFP-UVSSA. UVSS-A cells expressing free GFP were used as a control for non-specific binding proteins. All experiments were conducted in duplicates with a label swap and only proteins identified in both independent experiments (forward and reverse) with a log2 SILAC ratio of GFP-UVSSA/GFP above 0.6 were considered as specific UVSSA-interacting proteins. Results were visualized by plotting the log2 SILAC ratios of proteins of the two independent experiments (Figure 3A and Supplementary Table S1). In total, 66 specific UVSSA interactors were identified (Figure 3A, indicated in blue and orange). The bait UVSSA was identified with the highest SILAC ratio followed by USP7, a known UVSSA interactor (13,14,40), confirming the validity of our approach. To identify biological processes associated with these UVSSA-interacting proteins, we performed a Gene Ontology (GO) enrichment analysis. As expected, the biological process of DNA repair was among the top enriched GO annotations (Figure 3B). In addition, several of the top enriched GO terms were proteins involved in chromatin remodeling, suggesting that UVSSA interactors are involved in this process. Chromatin remodeling has been shown to play an important role in TC-NER (41–43), for example by enhancing the recruitment of TC-NER factors to DNA damage (44).
Figure 3.
Quantitative interaction proteomics reveal UVSSA interaction partners and required UVSSA-domains. (A) Scatter plot of log2 SILAC ratios of proteins isolated by GFP-pulldown in UVSS-A cells stably expressing either GFP-UVSSA or GFP (non-specific binding control). The experiment was conducted in duplicate with a label swap. The log2 SILAC ratios of proteins identified in the forward experiment (GFP-UVSSA versus GFP, H/L, x-axis) are plotted against the log2 SILAC ratios of proteins identified in the reversed experiment (GFP-UVSSA versus GFP, L/H, y-axis). Proteins were classified as specific UVSSA interactors (marked in blue) when log2 SILAC ratio >0.6 (indicated by gray dotted line) in both replicates. (B) GO-term analysis of the 66 proteins identified as specific interactors of UVSSA. A selection of the top 10 enriched biological process pathways is shown. GC: gene count; FDR: false discovery rate. (C) Scatter plot of log2 SILAC ratios of proteins identified in the GFP-pulldowns of wt UVSSA versus ΔVHS, only proteins that were also identified in the GFP-UVSSA versus GFP proteomics experiment are depicted. The experiment was conducted in duplicate, including a label swap. The log2 SILAC ratios of proteins identified in the forward experiment (wt versus ΔVHS, H/L, x-axis) are plotted against the log2 SILAC ratio of proteins identified in the reversed experiment (wt versus ΔVHS, L/H, y-axis). The majority of proteins have similar binding ability to the ΔVHS mutant compared to the wt (log2 SILAC ratio <0.6, proteins marked in gray). Proteins marked in blue represent proteins whose interaction with UVSSA is decreased in the absence of the VHS domain. (D) Scatter plot of log2 SILAC ratios of proteins identified in the GFP-pulldowns of wt UVSSA versus ΔDUF only proteins that were also identified in the GFP-UVSSA versus GFP proteomics experiment are depicted. The experiment was conducted in duplicate, including a label swap. The log2 SILAC ratios of proteins identified in the forward experiment (wt versus ΔDUF, H/L, x-axis) are plotted against the log2 SILAC ratio of proteins identified in the reversed experiment (wt versus ΔDUF, L/H, y-axis). Proteins marked in blue have a reduced interaction with UVSSAΔDUF compared to wt (proteins are marked in blue, log2 SILAC ratio >0.6, gray dotted line marks the threshold). (E) Cross-linked nuclear extracts of UVSS-A patient cell line (TA24), stably expressing the indicated constructs were subjected to GFP immunoprecipitation. Non-complemented UVSS-A patient cell line (-) was used as negative binding control. WCE: whole-cell extract, IP: Immunoprecipitation. Western blot analysis of the co-immunoprecipitated proteins was performed for GFP, Spt16, SSRP1, USP7 and CSA.
To pinpoint which proteins, from the 66 identified UVSSA interactors, are involved in the recruitment of UVSSA to DNA damage, we hypothesized that these proteins would retain their interaction with the ΔVHS mutant (the mutant that can localize to DNA damage), but might have lost binding with the ΔDUF mutant (the mutant that is not recruited to damage). Therefore, we performed quantitative proteomics experiments to map which of the identified UVSSA interactors are lost in the respective UVSSA deletion mutants (Supplementary Table S2). We compared proteins interacting with GFP-UVSSA (wt) to proteins interacting with GFP-UVSSA ΔVHS (Figure 3C) or GFP-UVSSA (wt) to GFP-UVSSA ΔDUF (Figure 3D). Log2 SILAC ratios around 0 indicate that proteins are equally bound to wt UVSSA as they are bound to the respective deletion mutant, while a positive log2 SILAC ratio indicates that the interaction with the tested deletion mutant is reduced compared to that with the wt UVSSA. Remarkably, only a few interactions were lost in the ΔVHS deletion mutant (Figure 3C, marked in blue), while numerous protein interactions with UVSSA were lost in the ΔDUF mutant (Figure 3D). This shows, in contrast to the interaction of the VHS domain with CSA (Figure 2C), that the majority of the identified UVSSA interactors depend on the DUF2043 domain.
Of the 66 proteins identified as UVSSA interactors (Figure 3A, indicated in blue), 45 proteins were detected in all three proteomic screens. The SILAC ratio of 25 of these proteins remained unchanged (SILAC ratio <1.2) in the ΔVHS mutant, indicating that the interactions between those proteins and UVSSA were similar for the wt UVSSA and the ΔVHS mutant. Since we hypothesized that proteins involved in the UVSSA recruitment to DNA damage would bind specifically to the DUF2043 domain (Figure 3D, indicated in blue), we sorted these remaining 25 proteins with descending wt/ΔDUF SILAC ratios (Supplementary Table S2). Interestingly, both the Spt16 and SSRP1 subunits of the histone chaperone complex FACT were identified as UVSSA interactors whose binding was lost most upon deletion of the DUF2043 domain.
FACT complex interaction with UVSSA is mediated by the DUF2043 domain
The H2A/H2B histone chaperone FACT is an interesting interaction partner of UVSSA, as it was originally discovered as an essential factor for productive in vitro Pol II transcription on chromatinized DNA (45) and plays essential roles in histone H2A/H2B exchange during DNA transcription and replication (46,47). Interestingly, recent studies have shown that FACT is involved in several DNA repair pathways (34,48–53). More specifically, Spt16 was shown to stimulate histone H2A/H2B exchange at sites of UV-induced DNA damage and to play an important role during the cellular response to TBLs by facilitating efficient restart of transcription following DNA damage removal (34). However, its exact mode of action and whether Spt16 is involved in TC-NER remains thus far unknown.
To test whether the role of Spt16 in transcription restart might be mediated via its identified interaction with UVSSA, we first confirmed the interaction between the FACT complex and UVSSA. Cross-linked immunoprecipitation experiments verified the interaction of the FACT complex with UVSSA and, as expected, with the known UVSSA interaction-partners CSA (27) and USP7 (13,14,40) (Figure 3E). This identified UVSSA–Spt16 interaction is strongly reduced when immunoprecipitation experiments are performed without cross-linking in the presence of benzonase, which degrades DNA and RNA (Supplementary Figure S3A). This indicates that the UVSSA interacts with Spt16 in a very transient manner, or that this interaction depends on the presence of RNA or DNA. In line with our proteomics data, we confirmed that FACT binding is mediated by the DUF2043 domain of UVSSA, as upon immunoprecipitation of the ΔDUF mutant Spt16 and SSRP1 could not be detected, whereas the interaction was present in the wt and ΔVHS mutant. Of note, the USP7 interaction with ΔDUF was significantly reduced, in agreement with our proteomics data (Figure 3D). However, the USP7 interaction was not completely lost, indicating that in addition to the DUF2043 domain, other UVSSA domains are also involved (40).
Spt16 enables efficient UVSSA recruitment and stimulates TC-NER-mediated damage excision
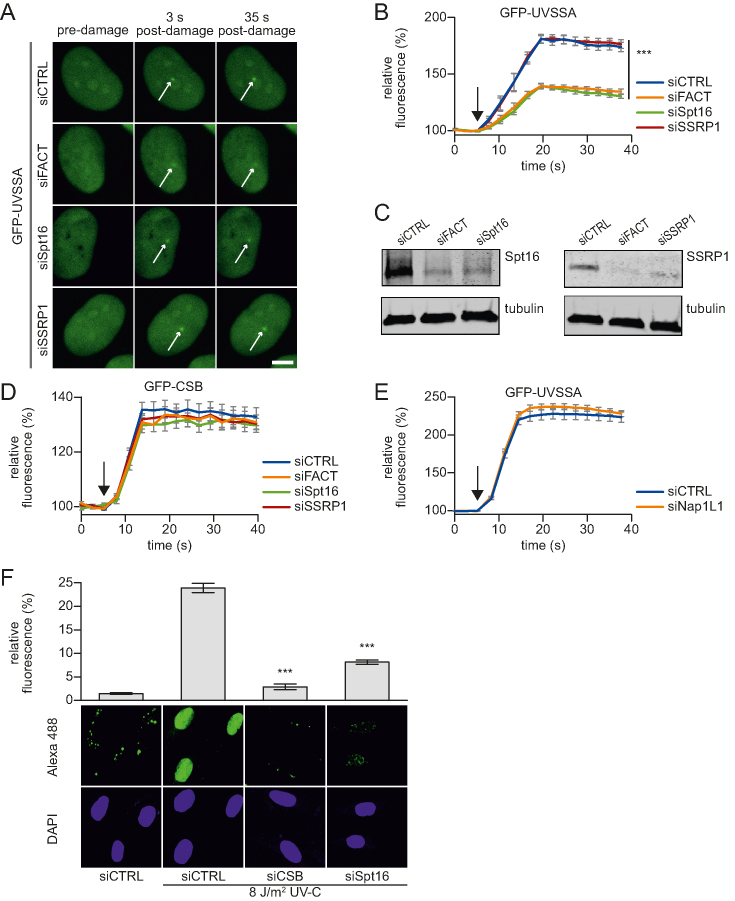
The identified UVSSA interaction with the FACT subunits SSRP1 and Spt16, together with the previously identified role of Spt16 in transcription restart, indicate that the FACT complex is involved in the function of UVSSA at sites of DNA damage. To test this, we quantified the UVSSA accumulation to sites of DNA damage in cells following siRNA-mediated knockdown of the FACT complex. Simultaneous knockdown of both FACT subunits significantly reduced the accumulation of UVSSA at LUD (Figure 4A and B). It is noteworthy that while knock down of Spt16 alone resulted in a comparable reduction of UVSSA accumulation, depletion of its canonical binding partner SSRP1, did not affect the accumulation of UVSSA, even though SSRP1 was efficiently depleted following siRNA transfection (Figure 4C). It should be noted, as is commonly observed for heterodimeric protein complexes, that siRNA-mediated knockdown of SSRP1 also results in the reduction of Spt16 protein levels, but not to the same extent as Spt16 knockdown itself (Supplementary Figure S3B). The remaining Spt16 protein levels after SSRP1 depletion are apparently sufficient to allow UVSSA recruitment. Furthermore, Spt16 knockdown also reduced the UVSSA accumulation in CSA-deficient patient cells (Supplementary Figure S3C and D), indicating that the residual UVSSA accumulation in TC-NER-proficient cells upon Spt16 depletion is not mediated by CSA.
Figure 4.
Spt16 mediates UVSSA accumulation on UV-C induced DNA damage. (A) Representative images of live-cell imaging analysis of GFP-UVSSA expressing cells transfected with the indicated siRNAs (CTRL is a non-targeting siRNA), following local UV-C laser (266 nm) induced damage (indicated by a white arrow); scale bar: 7.5 μm. (B) Relative GFP-UVSSA accumulation at sites of LUD in cells transfected with the indicated siRNA. GFP fluorescence intensity at LUD was measured over time using live-cell confocal imaging and normalized to pre-damage intensity set at 100 at t = 0 (n = 30 cells of two independent, pooled experiments, mean ± SEM). A one-way Anova test was performed and P-values <0.001 (***) are depicted. The moment of damage induction is indicated with a black arrow. (C) siRNA transfected cells as used in the live-cell imaging experiments (A and B) were lysed directly after the experiment. Lysates were analyzed by western blot with the indicated antibodies. Tubulin was used as loading control. (D) Relative GFP-CSB accumulation in CS-B (CS1AN) cells at sites of LUD in cells transfected with the indicated siRNA. GFP fluorescence intensity at LUD was measured over time using live-cell confocal imaging and normalized to pre-damage intensity set at 100 at t = 0 (n > 25 cells of two independent experiments, mean ± SEM). The black arrow indicated the moment of damage induction. Representative images are shown in Supplementary Figure S3E. (E) Relative GFP-UVSSA accumulation at sites of LUD in control and NAP1L1 depleted cells. Representative images and knock down efficacy are shown in Supplementary Figure S3F and G, respectively. GFP fluorescence intensity at LUD was measured over time using live-cell confocal imaging and normalized to pre-damage intensity set at 100 at t = 0 (n > 30 cells, two independent experiments, mean ± SEM). The black arrow indicates the moment of damage induction. (F) XP186LV patient cells (XP-C; GG-NER-deficient) were transfected with non-targeting control (CTRL) siRNA and siRNA against CSB and Spt16. Cells were irradiated with UV-C (8 J/m2) or mock-treated as indicated, and subsequently labelled for 7 h with EdU. The efficacy of the gap-filling synthesis was assessed by measuring the fluorescently labeled, incorporated EdU into the DNA. Amplified UDS signals were quantified (upper panel) by confocal microscopy measurement of the total nuclear fluorescence (Alexa-Fluor 488 nm, n > 170 cells for each condition, two independent experiments, mean ± SEM) and representative images (lower panel) are shown. A two-tailed t-test was performed and P-values < 0.001 (***) are depicted.
To test whether Spt16 depletion specifically affects UVSSA recruitment, or whether its absence inhibits the TC-NER complex assembly in general, we tested whether Spt16 depletion has a similar effect on CSB recruitment. In contrast to UVSSA, siRNA-mediated depletion of FACT did not affect CSB accumulation on UV-induced DNA damage (Figure 4D and Supplementary Figure S3E). As CSB is recruited to lesion-stalled Pol II (18), the absence of an effect of UVSSA on CSB recruitment suggests that the effect of Spt16 on UVSSA accumulation is not caused by a general effect on transcription or chromatin state. In line with this, the depletion of another H2A/H2B chaperone, the nucleosome assembly protein 1-like 1 (NAP1L1), did not interfere with UVSSA accumulation (Figure 4E; Supplementary Figure S3Fand G), indicating that UVSSA accumulation is not influenced by histone chaperones in general.
In accordance with the SSRP1-independent role of Spt16 in UVSSA recruitment, Spt16 was also shown to be specifically involved in transcription restart (34). This might suggest that the observed inhibition of transcription resumption is caused by a reduced UVSSA recruitment to sites of DNA damage, thereby inhibiting TC-NER efficiency and the subsequent transcription restart. To quantify TC-NER activity, we measured UDS by quantifying the DNA damage-induced EdU incorporation during gap-filling synthesis, which represents the last step of NER (36). To specifically quantify the TC-NER-mediated UDS, this assay was performed in non-cycling GG-NER-deficient cells (XP-C) and combined with a signal amplification step (36). In control siRNA-transfected cells, a clear UV-induced and TC-NER-specific UDS signal was observed, which was severely reduced following CSB depletion (Figure 4F). Interestingly, also upon siRNA-mediated knockdown of Spt16, the TC-NER-mediated UDS was significantly reduced. This indicates that Spt16 plays an important role in TC-NER by enhancing UVSSA recruitment, thereby subsequently stimulating transcription restart.
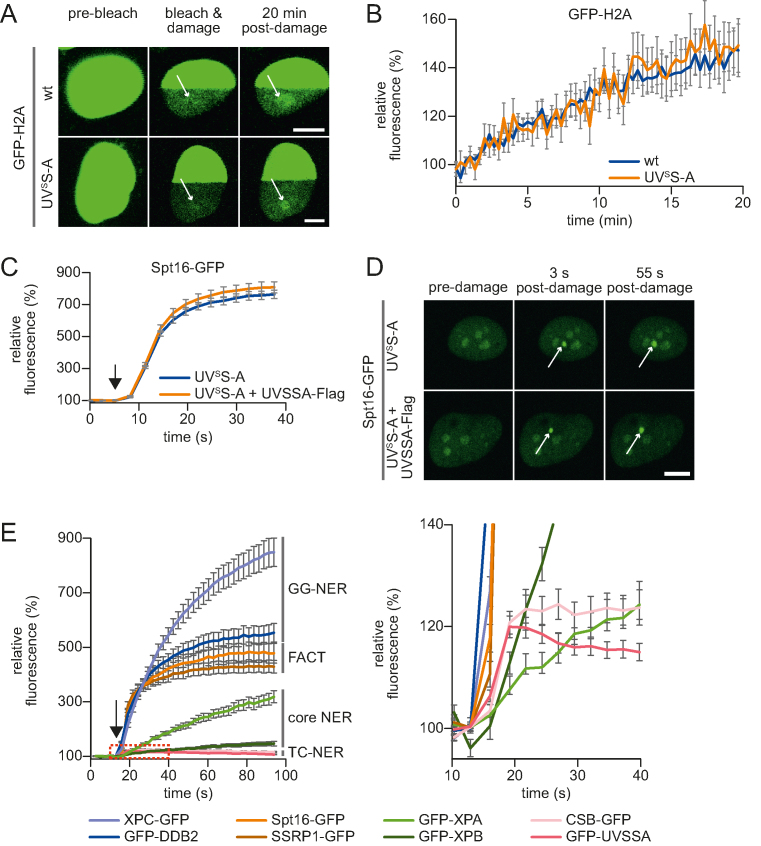
Spt16 function in TC-NER
Previously, we have shown that Spt16 stimulates accelerated exchange of histones H2A/H2B at sites of UV-induced damage (34). The identified interaction of Spt16 with UVSSA (Figure 3) together with the finding that both factors are recruited to DNA damage sites (14,34) (Figures 4A and 5D) prompted us to test whether UVSSA is involved in damage-induced accelerated exchange of histones H2A/H2B. Hence, we compared the UV-induced histone H2A exchange in UVSSA-proficient and -deficient cells. Histone exchange at DNA damage was determined in living cells by inducing local UV-C damage in a photobleached area of cells stably expressing GFP-H2A (34). Areas in the photobleached half of the nucleus that feature a higher histone exchange rate are characterized by an increased local GFP-H2A fluorescence due to eviction of photobleached GFP-H2A followed by incorporation of fluorescent (non-photobleached) GFP-H2A (Figure 5A). Histone exchange was quantified by comparing the recovery of GFP-H2A signal at sites of LUD with an undamaged region in the bleached part of the nucleus (Figure 5B). Both in UVSS-A (TA24) and TC-NER-proficient cells (HeLa), a comparable UV-induced H2A exchange was observed. This indicates that the Spt16-mediated H2A exchange at sites of DNA damage is independent of UVSSA. Together with the Spt16-dependent UVSSA accumulation, this suggests that the Spt16 recruitment acts in parallel or prior to UVSSA recruitment. To verify this, we compared the accumulation of stably expressed GFP-Spt16 to LUD in UVSS-A patient cells with UVSS-A complemented cells (Figure 5C and D; Supplementary Figure S4A). No difference could be observed in Spt16 accumulation. Similar results were obtained in U2OS cells stably expressing Spt16-GFP in which UVSSA was depleted using siRNA (Supplementary Figure S4B and C). The observation that Spt16 enhances the UVSSA accumulation to TBLs (Figure 4B), but not vice-versa, indicates that the activity of Spt16 is needed prior to the UVSSA recruitment during TC-NER.
Figure 5.
Spt16 is recruited to DNA damage early during TC-NER and independent of UVSSA. (A) Representative images of live-cell analysis of stable GFP-H2A expressing TC-NER proficient (HeLa cells, top panel) or UVSS-A (TA24 cells, lower panel) cells. Left panel, unbleached cells. After photobleaching half of the nucleus, (middle panel), local UV-damage (indicated with a white arrow) was inflicted with an UV-C (266 nm) laser in the bleached half of the nucleus. H2A-GFP exchange was imaged over time; scale bar: 7.5 μm. (B) The recovery of fluorescence in damaged and undamaged areas of the photobleached half of the nucleus is quantified in time. GFP-H2A exchange rate at LUD is depicted, normalized to the undamaged area (n = 15 cells from two independent mean ± SEM). (C) Relative Spt16-GFP accumulation at sites of LUD in UVSS-A deficient and complemented cells. GFP fluorescence intensity at LUD was measured over time using live-cell confocal imaging and was normalized to pre-damage intensity at t = 0, which was set to 100 (n = 30 cells, two independent experiments, mean ± SEM). The moment of damage induction is indicated with a black arrow. (D) Representative images of live-cell imaging analysis of Spt16-GFP expressed in either UVSS-A deficient or in UVSSA complemented (UVSSA-Flag) cells (lower panel). The white arrow indicates areas of UV-C laser (266 nm) induced DNA damage; scale bar: 7.5 μm. (E) Quantification of the GFP fluorescence intensity of cells stably expressing the indicated GFP-tagged NER proteins, GFP-tagged Spt16 or SSRP1. Cells subjected to local UV-C laser (266 nm) induced DNA damage were imaged over time. All cells were imaged and damaged under the exact same conditions. Fluorescence at LUD was normalized to pre-damage fluorescence at t = 0, which was set to 100 (n = 20 cells, two independent experiments, mean ± SEM). The moment of damage induction is indicated with a black arrow. Right graph shows zoomed graph of the indicated box, to clearly illustrate the kinetics of TC-NER factors. Representative images are shown in Supplementary Figure S4D.
To test whether Spt16 is indeed a factor that is recruited at the early stages of the TC-NER reaction, we directly compared its recruitment with the accumulation kinetics of a panel of GFP-tagged NER factors that are active at different stages in the NER reaction (Figure 5E and Supplementary Figure S4D). In line with previous studies (14,54), the DNA damage-recognizing GG-NER factors XPC and DDB2 showed a swift and robust accumulation (>6-fold change), while the core NER factors (XPA and XPB) involved in the more downstream damage verification step of NER featured slower accumulation kinetics with a more modest accumulation (1.5-to 3-fold change). The TC-NER-specific CSB and UVSSA proteins showed a quick but very modest accumulation (∼1.2-fold change). In line with a Spt16 recruitment upstream of UVSSA recruitment, Spt16 showed a very rapid accumulation. Remarkably, Spt16 showed a more pronounced accumulation (∼5-fold change) than UVSSA and CSB (∼1.2 fold) (Figure 5E), which might indicate a different mode of damage recruitment for Spt16 compared to TC-NER factors. Of note, even though not essential for UVSSA recruitment, SSRP1 showed similar accumulation as Spt16 (34).
Together, our data show that Spt16 functions as an early factor in the UV-DNA damage response (UV-DDR) and plays an important role in the recruitment of UVSSA, thereby stimulating efficient transcription-coupled repair and subsequent transcription resumption.
DISCUSSION
Recognition of lesion-stalled Pol II is a crucial initiating step for damage removal by TC-NER and its tight regulation is expected to be critical for correct spatiotemporal formation of the TC-NER complex, remodeling of lesion-stalled Pol II, and for the subsequent recruitment of downstream NER factors enabling efficient excision of TBLs (8,55). To gain further insight into this regulation, we investigated in detail the spatiotemporal behavior of the TC-NER initiating proteins CSA, CSB and UVSSA. We found that in living cells, the accumulation kinetics of CSB and UVSSA were strikingly similar, which might suggest similar modes of recruitment. Both proteins have been reported to have affinity for Pol II in unperturbed conditions, and their interaction is stabilized or their affinity is increased when bound to lesion-stalled Pol II (14,18,19). Despite their highly similar recruitment kinetics, UVSSA was recruited in a CSB-independent manner. Conversely, while CSB accumulation is reduced in the absence of UVSSA (13,14), this is most likely caused not by a direct effect of UVSSA on the initial CSB recruitment but rather by the stabilization of CSB via the deubiquitylating activity of the UVSSA binding partner USP7 (13,14). Interestingly, even though the crucial TC-NER factor CSA has been shown to directly interact with both UVSSA (27) and CSB (24), it could hardly be detected at sites of UV-induced DNA damage. This absence might be explained by the fact that CSA binds transiently to the TC-NER complex. This is in line with the general highly dynamic nature of interactions of E3 ligases with their substrates. Furthermore, this suggests that, in contrast to CSB and UVSSA, CSA has no structural or scaffold-like function in the TC-NER complex. Of note, the expected short residence time of CSA on TBLs makes it unlikely that CSA is responsible for recruiting UVSSA to UV-induced DNA damage via a direct protein interaction, as previously suggested based on cellular fractionation assays (13,27).
In this study, we precisely determined the accumulation kinetics of different TC-NER factors in living cells and show that UVSSA accumulation is similar in TC-NER-proficient and CSA- and CSB-deficient cells, indicating that the recruitment of UVSSA to sites of DNA damage is not influenced by CSA or CSB. The CSA-independent accumulation of UVSSA was further shown by the use of UVSSA deletion mutants lacking either the N-terminal VHS or the C-terminal DUF2043 domain. Of note, deletion of either domain resulted in a severe UV sensitivity and a reduced transcription resumption following irradiation (15). Interestingly, while deletion of the VHS domain resulted in the complete loss of CSA interaction (27), this mutant was recruited to DNA damage with exactly the same kinetics as full-length UVSSA. Conversely, deletion of the DUF2043 domain resulted in a severe reduction of UVSSA recruitment, without affecting the CSA interaction. These experiments show that the UVSSA recruitment at sites of TBLs can be separated from its interaction with CSA.
A plausible explanation for the apparent contradicting results on the role of CSA and CSB in the UVSSA recruitment (13,14,27,56) could be that the initial UVSSA recruitment to lesion-stalled Pol II, as determined in live-cell imaging experiments, is completely independent of CSA and CSB. However, the direct interaction between CSA and UVSSA, or another activity of CSA, might play an important role in the subsequent stabilization of the TC-NER complex. This complex-stabilizing function of CSA might explain the loss of UVSSA near TBLs in the absence of CSA (13,27), as weak or transient interactions might be lost during fractionation or immunoprecipitation assays. The CSA-independent UVSSA accumulation during live-cell experiments is also in line with previous studies. For example, the adenosine triphosphate (ATP)-dependent chromatin remodeler SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (SMARCA5) stimulates CSB (and presumably CSA) recruitment to UV-induced DNA damage, but does not affect UVSSA recruitment (44). Furthermore, while CSA is dispensable for the recruitment of TFIIH to lesion-stalled Pol II (57), functional TFIIH is essential for the CSA translocation to damaged chromatin (58). This data, together with the recent suggestion that UVSSA recruits TFIIH (25), supports a model in which UVSSA is recruited prior to and independent of CSA. This model raises an interesting question regarding the physiological function of the observed CSA–UVSSA interaction that can be detected even in unperturbed conditions (13,15,27,29). A specific mutation in CSA (W361C), which abolishes the interaction with UVSSA, results in the development of UVSS (27), indicating the importance of the CSA–UVSSA interaction for efficient TC-NER. It is tempting to speculate that the intrinsic affinity of UVSSA for CSA might be involved in the recruitment of the CRL4CSA E3 ligase to DNA damage. Otherwise, as CSA has affinity for CSB as well (24), it may play an important role in the stabilization or proper conformation of the TC-NER complex.
To identify proteins involved in the recruitment of UVSSA, we performed interaction proteomics. Among the top interacting proteins were the established UVSSA binding partners USP7 and the DDB1 and CUL4B subunits of the CRL4CSA complex (13,14,27), showing the validity of our approach (Figure 3A). To identify factors involved in UVSSA recruitment, we assumed that their interaction would be dependent on the DUF2043 domain, which is crucial for its localization to TBLs. Interestingly, in addition to the loss of interaction with the FACT subunits Spt16 and SSRP1, many other interactions were lost upon deletion of the DUF2043 domain, suggesting that this domain is a hotspot for interactions. For example, our MS analysis shows that the DUF2043 domain is essential for the UVSSA interaction with the U2 and U5 snRNP splicing factors SF3B1, SF3B2 and PRPF8. The interactions with these U2 and U5 snRNPs might be explained by the affinity of UVSSA for elongating Pol II in both unperturbed or DNA damage conditions (14). These late-stage splicing factors have been shown to be displaced from the chromatin following TBL induction, thereby increasing R-loop formation and activation of ATM signaling (59). The identified interaction with these splicing factors might indicate a role for UVSSA in these transcription-coupled processes during the DNA damage response.
In this study, we focused on the role of Spt16 in the regulation of UVSSA and TC-NER, as we have previously identified this subunit of the H2A/H2B histone chaperone FACT to be involved in the UV-induced H2A/H2B exchange and to stimulate transcription restart following DNA damage (34). However, thus far, the exact mechanism of how Spt16 regulates TC-NER remains unknown. In line with the interaction of Spt16 being dependent on the DUF2043 domain of UVSSA, we found that Spt16 is required for the recruitment of UVSSA to damaged DNA. The ∼50% reduction of UVSSA recruitment following SPT16 depletion, caused severe effects on the TC-NER-mediated repair, as shown by a strong impediment of the TC-NER-specific gap-filling synthesis. This reduced repair efficiency can explain the previously observed inhibition of transcription restart and UV sensitivity upon Spt16 knockdown (34). However, additional effects of Spt16 on the transcription restart process independent of repair, as shown for Dot1L (60), cannot be excluded. As Spt16 depletion (Figure 4B), or deletion of the DUF2043 domain (Figure 2E), does not result in a full loss of UVSSA recruitment at sites of DNA damage, it is likely that additional mechanisms and factors are involved in the recruitment of UVSSA.
The effects of Spt16 on UVSSA recruitment, as well as transcription restart and UV sensitivity (34), are independent of SSRP1, its canonical binding partner in the FACT complex. Despite being not essential for TC-NER, SSRP1 interacts with UVSSA and has similar accumulation kinetics at DNA damage sites as Spt16. Spt16 seems to be the driving force of the FACT accumulation, as the SSRP1 accumulation at UV-induced DNA damage depends on the presence of Spt16 (34). Together, this suggests that under normal conditions, the complete FACT heterodimer is present at sites of DNA damage, but in the absence of SSRP1, Spt16 can be recruited and function during TC-NER independently of SSRP1. Although, thus far, it remains unknown how Spt16 and SSRP1 are exactly recruited to sites of DNA damage, Spt16 accumulation occurs early during the repair reaction and independently of other TC-NER-initiating factors (Figure 5C–E) (34). In addition, Spt16 and SSRP1 showed, in comparative DNA damage accumulation experiments, a striking pattern. Similar to TC-NER factors, FACT accumulated almost instantaneously following DNA damage infliction. However, while CSB and UVSSA only showed a modest ∼1.2-fold accumulation, FACT subunits showed a 5-fold accumulation, almost to the same extent as the highly efficient DNA damage-recognizing GG-NER factors DDB2 and XPC. This might suggest that Spt16 has, in addition to what is described for SSRP1 (50,53), damage-recognizing capabilities, either by directly recognizing the lesion, or indirectly, for example by sensing damage-induced transcription impediment. Furthermore, these differences in fold accumulation might indicate that a multitude of Spt16 molecules are present near TBLs compared to the TC-NER proteins CSB and UVSSA.
We found that H2A/H2B exchange at sites of DNA damage was independent of UVSSA. This indicates that UVSSA interaction with Spt16 is not needed to induce accelerated histone exchange at TBLs, but rather suggests that Spt16-mediated histone exchange mediates efficient UVSSA recruitment or, alternatively, that it is important for the stable incorporation of UVSSA in the TC-NER complex. However, this effect is not caused by a general inhibition of histone turnover, as knockdown of NAP1L1, another H2A/H2B chaperone, did not affect UVSSA recruitment (Figure 4E).
Although transcription and thus also TC-NER, occurs in a more open and therefore accessible chromatin state, several chromatin-remodeling enzymes were shown to be necessary for efficient repair and transcription restart (4,42). For example, Nap1L1 stimulates the ATP-dependent chromatin-remodeling activity of CSB (61). In addition, in our UVSSA interaction screen, we also identified CHD4 as a putative TC-NER-involved chromatin remodeler (Supplementary Table S1). While CHD4 has been reported to be involved in DDR (62,63), it is currently unknown whether it is involved in TC-NER. Thus far, the only two chromatin-modifying factors shown to influence accumulation of TC-NER factors are Spt16, which stimulates UVSSA recruitment, and the ATP-dependent chromatin remodeler SMARCA5, which facilitates recruitment of CSB (44). Interestingly, both factors act at specific TC-NER reaction steps; SMARCA5 does not affect UVSSA recruitment and Spt16 is not involved in the CSB recruitment. This suggests that the involvement of these chromatin modifiers in TC-NER is not restricted simply to making the chromatin accessible. The need for different chromatin-modifying enzymes for recruitment of CSB and UVSSA strengthens our observation that despite their similar accumulation kinetics, these TC-NER-initiating factors are independently recruited (14) to damaged chromatin. Furthermore, this suggests that both SMARCA5 and Spt16 stimulate specific changes in the chromatin, for example, nucleosome sliding or histone exchange, that are important during different TC-NER reaction steps. In line with this notion, distinct functions for CSB and UVSSA during TC-NER have been described. CSB was suggested to stimulate Pol II forward translocation, analogous to the action of Mfd in prokaryotes (64), thereby discriminating between lesion-stalled Pol II and other non-forward translocating Pol II complexes, for example, Pol II stalled on naturally occurring pause sites (19). UVSSA was shown to recruit TFIIH via a direct interaction with P62 in a similar manner as XPC recruits TFIIH in GG-NER (65). Collectively, these observations would suggest that SMARCA5 is involved in remodeling lesion-stalled Pol II, while Spt16 either recruits or allows TFIIH to properly function during the TC-NER reaction. In summary, this study provides important new insight into the regulation of TC-NER and, more specifically, into the assembly of the TC-NER complex. Furthermore, these results highlight that different chromatin-modulating factors regulate distinct steps of the highly orchestrated TC-NER pathway.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Optical Imaging Centre (OIC) of the Erasmus MC for support with microscopes and Dr Arjan Theil for help with FACS sorting.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Dutch organization for Scientific Research ZonMW TOP Grant [912.12.132]; Horizon Zenith [935.11.042]; Dutch organization for Scientific Research (NWO-ALW) VIDI [864.13.004]; Erasmus MC fellowship. Thiswork is part of the Oncode Institute which is partly financed by the Dutch Cancer Society and was funded by a grant from the Dutch Cancer Society [KWF 10506].
Conflict of interest statement. None declared.
REFERENCES
- 1. Brueckner F., Hennecke U., Carell T., Cramer P.. CPD damage recognition by transcribing RNA polymerase II. Science. 2007; 315:859–862. [DOI] [PubMed] [Google Scholar]
- 2. Shin J.H., Xu L., Wang D.. RNA polymerase II acts as a selective sensor for DNA lesions and endogenous DNA modifications. Transcription. 2016; 7:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vermeij W.P., Hoeijmakers J.H., Pothof J.. Aging: not all DNA damage is equal. Curr. Opin. Genet. Dev. 2014; 26:124–130. [DOI] [PubMed] [Google Scholar]
- 4. Steurer B., Marteijn J.A.. Traveling rocky Roads: the consequences of transcription-blocking DNA lesions on RNA polymerase II. J. Mol. Biol. 2017; 429:3146–3155. [DOI] [PubMed] [Google Scholar]
- 5. Hanawalt P.C., Spivak G.. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008; 9:958–970. [DOI] [PubMed] [Google Scholar]
- 6. Marteijn J.A., Lans H., Vermeulen W., Hoeijmakers J.H.. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014; 15:465–481. [DOI] [PubMed] [Google Scholar]
- 7. Tornaletti S. Transcription arrest at DNA damage sites. Mutation Res. 2005; 577:131–145. [DOI] [PubMed] [Google Scholar]
- 8. Spivak G. Transcription-coupled repair: an update. Arch. Toxicol. 2016; 90:2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spivak G. UV-sensitive syndrome. Mutation Res. 2005; 577:162–169. [DOI] [PubMed] [Google Scholar]
- 10. Karikkineth A.C., Scheibye-Knudsen M., Fivenson E., Croteau D.L., Bohr V.A.. Cockayne syndrome: clinical features, model systems and pathways. Ageing Res. Rev. 2017; 33:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Laugel V. Cockayne syndrome: the expanding clinical and mutational spectrum. Mech. Ageing Dev. 2013; 134:161–170. [DOI] [PubMed] [Google Scholar]
- 12. Cleaver J.E., Lam E.T., Revet I.. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 2009; 10:756–768. [DOI] [PubMed] [Google Scholar]
- 13. Zhang X., Horibata K., Saijo M., Ishigami C., Ukai A., Kanno S., Tahara H., Neilan E.G., Honma M., Nohmi T. et al.. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat. Genet. 2012; 44:593–597. [DOI] [PubMed] [Google Scholar]
- 14. Schwertman P., Lagarou A., Dekkers D.H., Raams A., van der Hoek A.C., Laffeber C., Hoeijmakers J.H., Demmers J.A., Fousteri M., Vermeulen W. et al.. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat. Genet. 2012; 44:598–602. [DOI] [PubMed] [Google Scholar]
- 15. Nakazawa Y., Sasaki K., Mitsutake N., Matsuse M., Shimada M., Nardo T., Takahashi Y., Ohyama K., Ito K., Mishima H. et al.. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat. Genet. 2012; 44:586–592. [DOI] [PubMed] [Google Scholar]
- 16. Spivak G. The many faces of Cockayne syndrome. Proc. Nat. Acad. Sci. U.S.A. 2004; 101:15273–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Selby C.P., Sancar A.. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 1997; 272:1885–1890. [DOI] [PubMed] [Google Scholar]
- 18. van den Boom V., Citterio E., Hoogstraten D., Zotter A., Egly J.M., van Cappellen W.A., Hoeijmakers J.H., Houtsmuller A.B., Vermeulen W.. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J. Cell. Biol. 2004; 166:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu J., Lahiri I., Wang W., Wier A., Cianfrocco M.A., Chong J., Hare A.A., Dervan P.B., DiMaio F., Leschziner A.E. et al.. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017; 551:653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J.H.. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992; 71:939–953. [DOI] [PubMed] [Google Scholar]
- 21. Kamiuchi S., Saijo M., Citterio E., de Jager M., Hoeijmakers J.H., Tanaka K.. Translocation of Cockayne syndrome group A protein to the nuclear matrix: possible relevance to transcription-coupled DNA repair. Proc. Nat. Acad. Sci. U.S.A. 2002; 99:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A.F., Tanaka K., Nakatani Y.. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003; 113:357–367. [DOI] [PubMed] [Google Scholar]
- 23. Fischer E.S., Scrima A., Bohm K., Matsumoto S., Lingaraju G.M., Faty M., Yasuda T., Cavadini S., Wakasugi M., Hanaoka F. et al.. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011; 147:1024–1039. [DOI] [PubMed] [Google Scholar]
- 24. Groisman R., Kuraoka I., Chevallier O., Gaye N., Magnaldo T., Tanaka K., Kisselev A.F., Harel-Bellan A., Nakatani Y.. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 2006; 20:1429–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okuda M., Higo J., Komatsu T., Konuma T., Sugase K., Nishimura Y. Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways. Nucleic Acids Res. 2017; 45:13043–13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Boer J., Hoeijmakers J.H.. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000; 21:453–460. [DOI] [PubMed] [Google Scholar]
- 27. Fei J., Chen J.. KIAA1530 protein is recruited by Cockayne syndrome complementation group protein A (CSA) to participate in transcription-coupled repair (TCR). J. Biol. Chem. 2012; 287:35118–35126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Campeau E., Ruhl V.E., Rodier F., Smith C.L., Rahmberg B.L., Fuss J.O., Campisi J., Yaswen P., Cooper P.K., Kaufman P.D.. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009; 4:e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pines A., Dijk M., Makowski M., Meulenbroek E.M., Vrouwe M.G., van der Weegen Y., Baltissen M., French P.J., van Royen M.E., Luijsterburg M.S. et al.. TRiC controls transcription resumption after UV damage by regulating Cockayne syndrome protein A. Nat. Commun. 2018; 9:1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoogstraten D., Bergink S., Ng J.M., Verbiest V.H., Luijsterburg M.S., Geverts B., Raams A., Dinant C., Hoeijmakers J.H., Vermeulen W. et al.. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J. Cell Sci. 2008; 121:2850–2859. [DOI] [PubMed] [Google Scholar]
- 31. Rademakers S., Volker M., Hoogstraten D., Nigg A.L., Mone M.J., Van Zeeland A.A., Hoeijmakers J.H., Houtsmuller A.B., Vermeulen W.. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol. Cell. Biol. 2003; 23:5755–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoogstraten D., Nigg A.L., Heath H., Mullenders L.H.F., van Driel R., Hoeijmakers J.H.J., Vermeulen W., Houtsmuller A.B.. Rapid switching of TFIIH between RNA Polymerase I and II transcription and DNA repair in vivo. Mol. Cell. 2002; 10:1163–1174. [DOI] [PubMed] [Google Scholar]
- 33. Pines A., Vrouwe M.G., Marteijn J.A., Typas D., Luijsterburg M.S., Cansoy M., Hensbergen P., Deelder A., de Groot A., Matsumoto S. et al.. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 2012; 199:235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dinant C., Ampatziadis-Michailidis G., Lans H., Tresini M., Lagarou A., Grosbart M., Theil A.F., van Cappellen W.A., Kimura H., Bartek J. et al.. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol. Cell. 2013; 51:469–479. [DOI] [PubMed] [Google Scholar]
- 35. Dinant C., de Jager M., Essers J., van Cappellen W.A., Kanaar R., Houtsmuller A.B., Vermeulen W.. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 2007; 120:2731–2740. [DOI] [PubMed] [Google Scholar]
- 36. Wienholz F., Vermeulen W., Marteijn J.A.. Amplification of unscheduled DNA synthesis signal enables fluorescence-based single cell quantification of transcription-coupled nucleotide excision repair. Nucleic Acids Res. 2017; 45:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M.. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011; 10:1794–1805. [DOI] [PubMed] [Google Scholar]
- 39. Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., Leonhardt H.. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 2008; 7:282–289. [DOI] [PubMed] [Google Scholar]
- 40. Higa M., Zhang X., Tanaka K., Saijo M.. Stabilization of ultraviolet (UV)-stimulated scaffold protein A by interaction with Ubiquitin-specific peptidase 7 is essential for Transcription-coupled nucleotide excision repair. J. Bio.l Chem. 2016; 291:13771–13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lans H., Marteijn J.A., Vermeulen W.. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 2012; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mandemaker I.K., Vermeulen W., Marteijn J.A.. Gearing up chromatin: a role for chromatin remodeling during the transcriptional restart upon DNA damage. Nucleus. 2014; 5:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Czaja W., Mao P., Smerdon M.J.. The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair. Int. J. Mol. Sci. 2012; 13:11954–11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aydin O.Z., Marteijn J.A., Ribeiro-Silva C., Rodriguez Lopez A., Wijgers N., Smeenk G., van Attikum H., Poot R.A., Vermeulen W., Lans H.. Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription. Nucleic Acids Res. 2014; 42:8473–8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Orphanides G., LeRoy G., Chang C.H., Luse D.S., Reinberg D.. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998; 92:105–116. [DOI] [PubMed] [Google Scholar]
- 46. Tsunaka Y., Fujiwara Y., Oyama T., Hirose S., Morikawa K.. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 2016; 30:673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Winkler D.D., Luger K.. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011; 286:18369–18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao Y., Li C., Wei L., Teng Y., Nakajima S., Chen X., Xu J., Legar B., Ma H., Spagnol S.T. et al.. SSRP1 cooperates with PARP and XRCC1 to facilitate single-strand DNA break repair by chromatin priming. Cancer Res. 2017; 77:2674–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Charles Richard J.L., Shukla M.S., Menoni H., Ouararhni K., Lone I.N., Roulland Y., Papin C., Ben Simon E., Kundu T., Hamiche A. et al.. FACT assists base excision repair by boosting the remodeling activity of RSC. PLoS Genet. 2016; 12:e1006221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yarnell A.T., Oh S., Reinberg D., Lippard S.J.. Interaction of FACT, SSRP1, and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J. Biol. Chem. 2001; 276:25736–25741. [DOI] [PubMed] [Google Scholar]
- 51. Oliveira D.V., Kato A., Nakamura K., Ikura T., Okada M., Kobayashi J., Yanagihara H., Saito Y., Tauchi H., Komatsu K.. Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J. Cell Sci. 2014; 127:763–772. [DOI] [PubMed] [Google Scholar]
- 52. Keller D.M., Lu H.. p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J. Biol. Chem. 2002; 277:50206–50213. [DOI] [PubMed] [Google Scholar]
- 53. Krohn N.M., Stemmer C., Fojan P., Grimm R., Grasser K.D.. Protein kinase CK2 phosphorylates the high mobility group domain protein SSRP1, inducing the recognition of UV-damaged DNA. J. Biol. Chem. 2003; 278:12710–12715. [DOI] [PubMed] [Google Scholar]
- 54. Marteijn J.A., Bekker-Jensen S., Mailand N., Lans H., Schwertman P., Gourdin A.M., Dantuma N.P., Lukas J., Vermeulen W.. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 2009; 186:835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Marteijn J.A., Hoeijmakers J.H., Vermeulen W.. Check, check …triple check: multi-step DNA lesion identification by nucleotide excision repair. Mol. Cell. 2015; 59:885–886. [DOI] [PubMed] [Google Scholar]
- 56. Schwertman P., Vermeulen W., Marteijn J.A.. UVSSA and USP7, a new couple in transcription-coupled DNA repair. Chromosoma. 2013; 122:275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fousteri M., Mullenders L.H.. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008; 18:73–84. [DOI] [PubMed] [Google Scholar]
- 58. Saijo M., Hirai T., Ogawa A., Kobayashi A., Kamiuchi S., Tanaka K.. Functional TFIIH is required for UV-induced translocation of CSA to the nuclear matrix. Mol. Cell. Biol. 2007; 27:2538–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tresini M., Warmerdam D.O., Kolovos P., Snijder L., Vrouwe M.G., Demmers J.A., van I.W.F., Grosveld F.G., Medema R.H., Hoeijmakers J.H. et al.. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015; 523:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oksenych V., Zhovmer A., Ziani S., Mari P.O., Eberova J., Nardo T., Stefanini M., Giglia-Mari G., Egly J.M., Coin F.. Histone methyltransferase DOT1L drives recovery of gene expression after a genotoxic attack. PLoS Genet. 2013; 9:e1003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cho I., Tsai P.F., Lake R.J., Basheer A., Fan H.Y.. ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS Genet. 2013; 9:e1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Burd C.J., Kinyamu H.K., Miller F.W., Archer T.K.. UV radiation regulates Mi-2 through protein translation and stability. J. Biol. Chem. 2008; 283:34976–34982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. O'Shaughnessy A., Hendrich B.. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem. Soc. Trans. 2013; 41:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pani B., Nudler E.. Mechanistic insights into transcription coupled DNA repair. DNA Repair. 2017; 56:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Okuda M., Nakazawa Y., Guo C., Ogi T., Nishimura Y.. Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways. Nucleic Acids Res. 2017; 45:13043–13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.