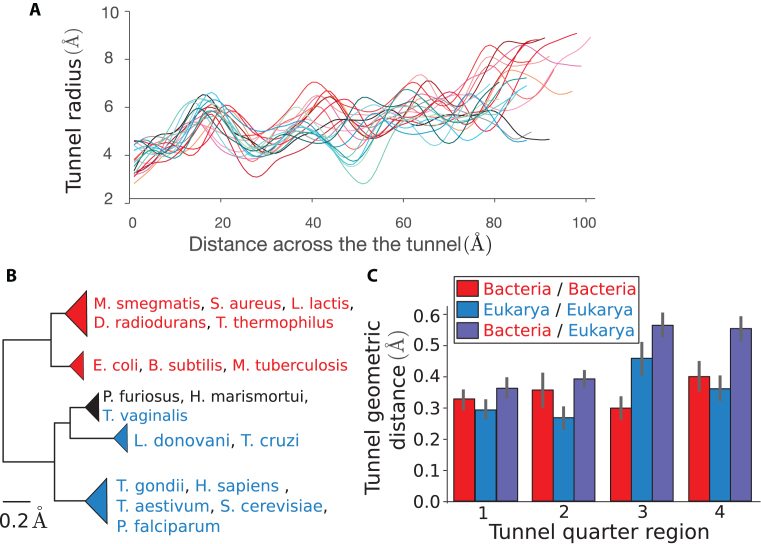
Figure 3.
Clustering of species obtained from pairwise comparison of the tunnel geometry. (A) For all our structures, we plot the tunnel radius as a function of the distance across the tunnel. These plots are used to compare the tunnel geometries. (B) Clustering obtained after applying our tunnel geometric distance metric to our dataset (for a definition of the metric and more details, see ‘Materials and methods’ section). The first main branch encompasses the bacterial ribosomes, highlighted in red, while the second contains eukarya (blue) and archaea (black) (scale bar: 0.2 Å). For the full clustering and phylogenetic trees obtained from 16S/18S rRNA sequences, see Supplementary Figure S2. (C) We divide for each couple of species their common domain after alignment in 4 quarters (see ‘Materials and methods’ section) and use the same metric to compute the distance in each of the subregions. The bar plots represent for each quarter the average and std of the geometric distance for subset of pairs made of 2 prokaryotes (red), 2 eukaryotes (blue), and 1 prokaryote and 1 eukaryote (violet).