Figure 3.
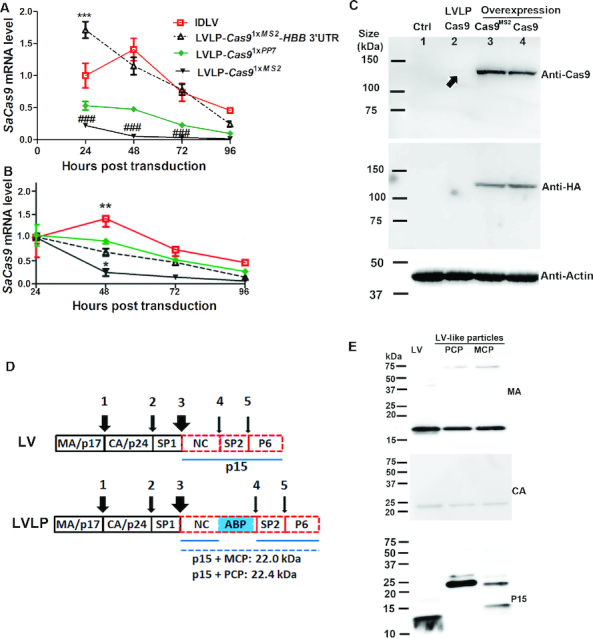
Characterization of LVLPs. (A) Transient expression of SaCas9 mRNA from LVLPs. SaCas9-expressing IDLV (35 ng p24) or LVLPs (35 ng p24) were transduced into 2.5 × 104 HEK293T cells. SaCas9 mRNA levels were assayed at different time points. SaCas9 mRNA levels 24h post transduction were normalized by housekeeping gene GAPDH and RPLP0 to obtain the relative SaCas9 mRNA expression level. No normalization was performed thereafter so that possible cell replication does not affect evaluation of mRNA degradation since no new mRNA was generated in LVLP transduced cells. *** indicates P < 0.001 when SaCas91xMS2-HBB 3′UTR was compared with other particles at the same time. ### indicates P < 0.001 when SaCas91xMS2 was compared with other particles at the same time. (B) Time course of SaCas9 mRNA level from the same particle. mRNA levels of each particle 24h post transduction were set as 1. Shown are mean ± SEM of indicated replicates. * and ** indicate P < 0.05 and P < 0.01 when compared with other particles at the same time point. For A and B, Bonferroni posttests were performed following two-way ANOVA. (C) Western blotting analysis of SaCas9 protein. The four lanes were lysates from mock transfected HREK293T cells (lane 1), GFP-reporter cells co-transduced with 300 ng p24 of Cas9 LVLP and 50 ng p24 of HBB sgRNA1 IDLV (lane 2), HEK293T cells overexpressing Cas91xMS2 (lane 3) or Cas9 mRNA (lane 4) by transfecting 0.25 μg DNA to 1.25 × 105 cells. A very faint band in LVLP transduced cells was indicated by an arrow. (D) Diagram showing the processing of Gag precursor by HIV protease. The wideness of the arrows is proportional to the processing speed at that site (1–5). The estimated sizes of the p15-ABP fusion proteins were listed. (E) Western blotting analysis of lentiviral proteins. 200 ng p24 of GFP lentivirus, NC-MCP and NC-PCP modified LVLPs were analyzed.
