Figure 4.
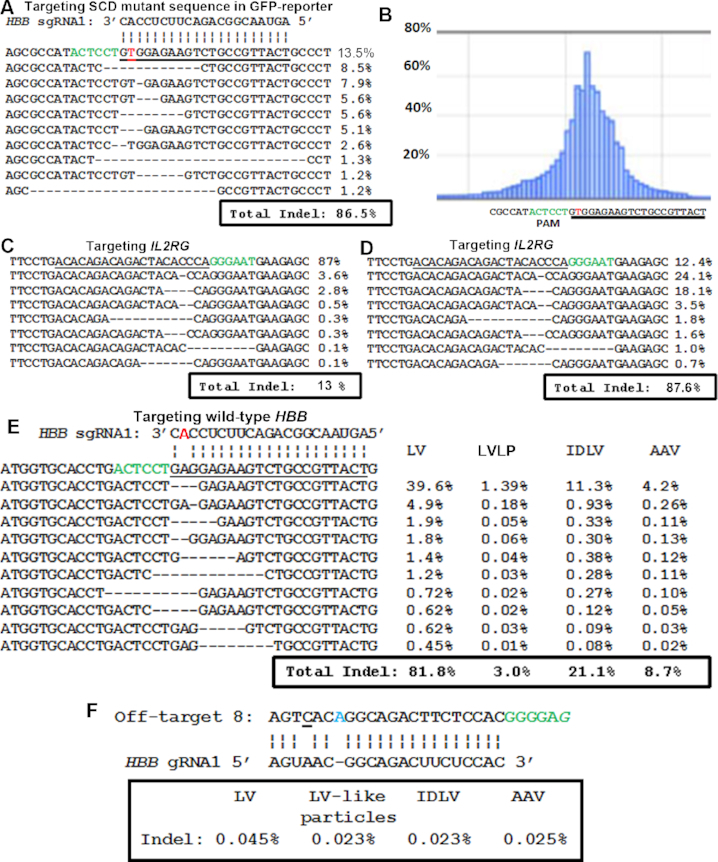
Genome editing by SaCas9 LVLP. (A) SaCas9 LVLPs efficiently generated Indels in the perfectly matched HBB sgRNA1 target sequence. The most frequently observed sequences and their percentages from next-generation sequencing are listed. The ‘T’ in red is the mutation in HBB causing Sickle cell disease. (B) Frequency of Indels at each position obtained from sequencing data in A. (C) Indels in IL2RG gene of HEK293T cells generated by SaCas9 LVLPs. 2.5 × 104 cells were co-transduced with 30 ng p24 of SaCas91xMS2-HBB 3′UTR LVLPs and 60 ng of IDLV expressing IL2RG sgRNA. (D) Indels in IL2RG gene of lymphoblasts generated by 500 ng p24 of SaCas91xMS2-HBB 3′UTR LVLPs (500 ng p24 LVLPs on 2 × 105 cells). (E) Indel rates in the wild-type HBB locus of the GFP reporter cells. There was one mismatch between the HBB sgRNA1 (highlighted in red) and the target sequence. (F) Indel rates in predicted off-target 8 of the GFP reporter cells. Off-target 8 has one mismatch with HBB sgRNA1 (underlined), one DNA bulge (in blue) and a non-typical PAM (a ‘G’ instead of a ‘T’ at the last position, in italics). For A–F: The protospacer adjacent motif (PAM) or its complementary sequence is in green. For A–E: The target sequences are underlined. Vertical lines indicate complementarity between HBB sgRNA1 and the target. 3–4 million readings were obtained for each sample.
