Figure 5.
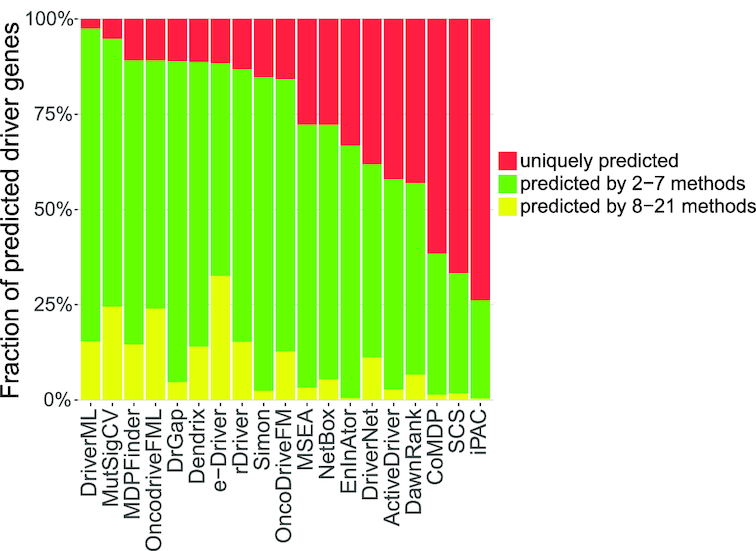
Fraction of predicted driver genes for each method by consensus among the methods. The average fraction of predicted driver genes for each method was determined by consensus among the other methods for the 31 datasets. Tools were sorted by the fraction of uniquely predicted drivers (indicated in red) from small to large. OncodriveCLUST was removed because it did not predicted any unique driver genes on 31 data sets. MEMo was also removed because it predicted too few genes. DriverML, MutSigCV and MDPFinder had the smallest fractions of uniquely predicted driver genes (2.5%, 5.2% and 10.8%, respectively). iPAC, SCS and CoMDP had the highest fractions (73.8%, 66.7% and 61.6%, respectively).
