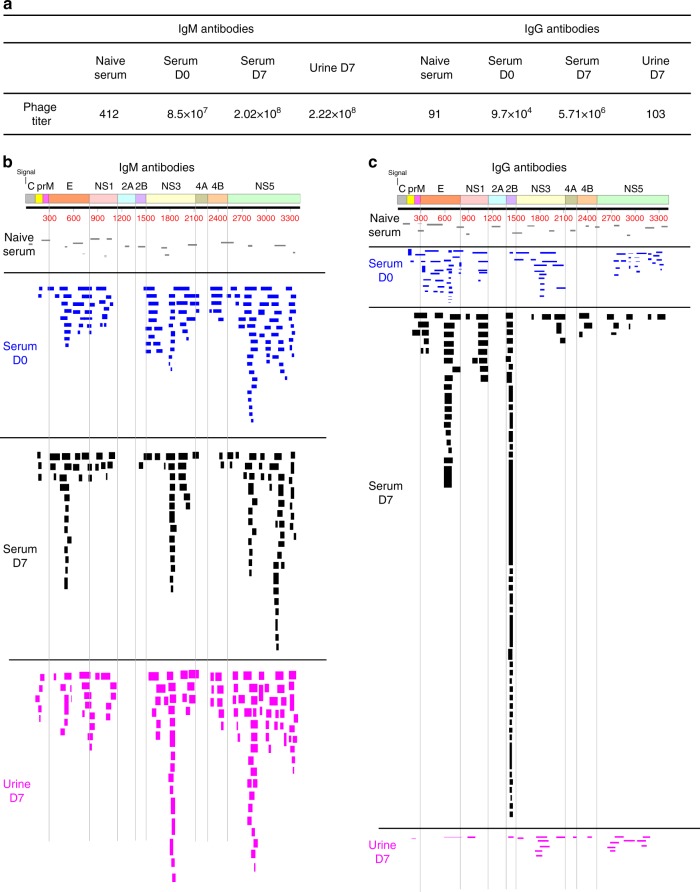
Fig. 1.
IgG and IgM repertoires across Zika virus (ZIKV) proteome following Zika virus infection. a Number of bound phage clones isolated using ZIKV GFPDL affinity selection against flavivirus naive negative control serum, pooled serum samples from five acutely ZIKV-infected patients on day 0 and day 7 of hospital visit, and urine sample on day 7 of hospital visit (day 0 of hospital visit corresponds to 0–3 days since onset of symptoms for these five patients). b, c Schematic alignment of the peptide sequences recognized by IgM (b) and IgG (c) antibodies in ZIKV-infected human sera (day 0 and day 7) and urine (day 7) and flavivirus naive negative control serum, identified by panning with ZIKV-GFPDL. The amino-acid designation is based on the ZIKV polyprotein sequence encoded by the complete ZIKV-ICD genome (Supplementary Fig. 1). Bars indicate identified epitopes in the different structural (C, prM, E) and non-structural (NS) genes on the ZIKV polyprotein sequence. Graphical distribution of representative clones with a frequency of >2, obtained after affinity selection, are shown. The horizontal position and the length of the bars indicate the peptide sequence displayed on the selected phage clone to its homologous sequence in the ZIKV sequence on alignment. The thickness of each bar represents the frequency of repetitively isolated phages. Phage sequences and clonal frequency is shown in Supplementary Table 3