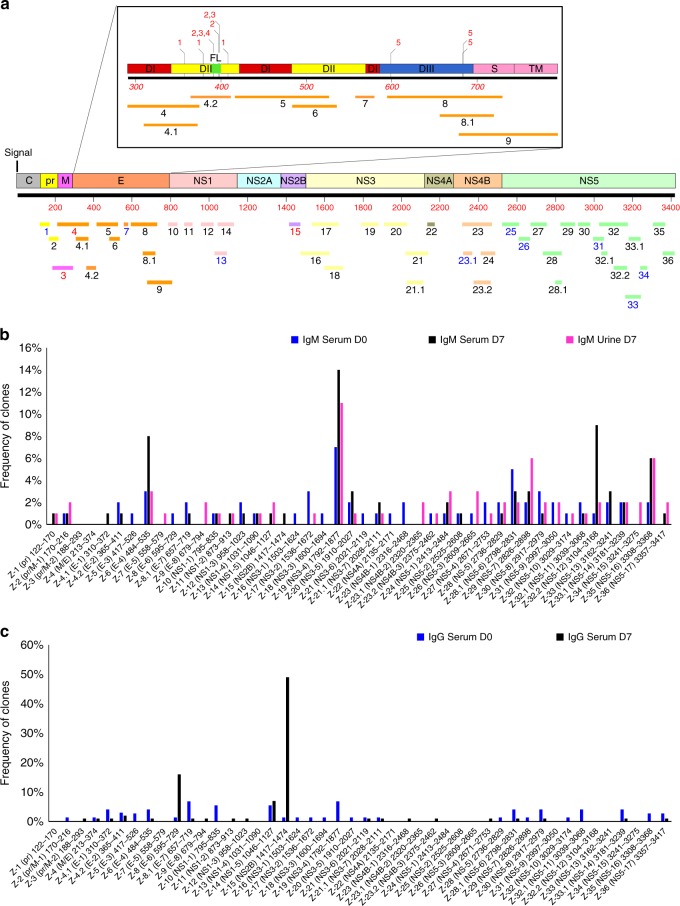
Fig. 2.
Frequency of Zika virus (ZIKV) epitopes in serum and urine following Zika virus infection. a Antigenic sites within the ZIKV proteins recognized by serum (days 0, 7) and urine (day 7) IgM and IgG antibodies following Zika virus infection (based on data presented in Fig. 1). The amino-acid designation is based on the ZIKV polyprotein sequence encoded by the complete ZIKV genome (Supplementary Fig. 1). The amino-acid designations on the ZIKV polyprotein (Supplementary Fig. 1) are as follows: C capsid, pr peptide pr, M membrane, E envelope protein, NS non-structural protein. Inset shows expanded version of E protein schematic with domains (D) I, II, and III and fusion loop (FL) shown along with their antigenic sites. Previously described epitopes using MAbs are shown above the ZIKV-E schematic. Critical residues for binding of MAbs, 1, ZIKV-117; 2, 2A10G6; 3, ZIKV-12; 4, ZIKV-15; and 5, ZIKV-116 are depicted. b, c Distribution and frequency of phage clones expressing each of the key ZIKV antigenic sites recognized by IgM (b) and IgG (c) antibodies in post-ZIKV infection serum (day 0 and day 7) and urine (day 7) are shown. The number of phage clones that expressed each antigenic site was divided by the total number of ZIKV-GFPDL selected clones for each pooled sera or urine samples and represented as a percentage. Phage sequences and clonal frequency is shown in Supplementary Table 3