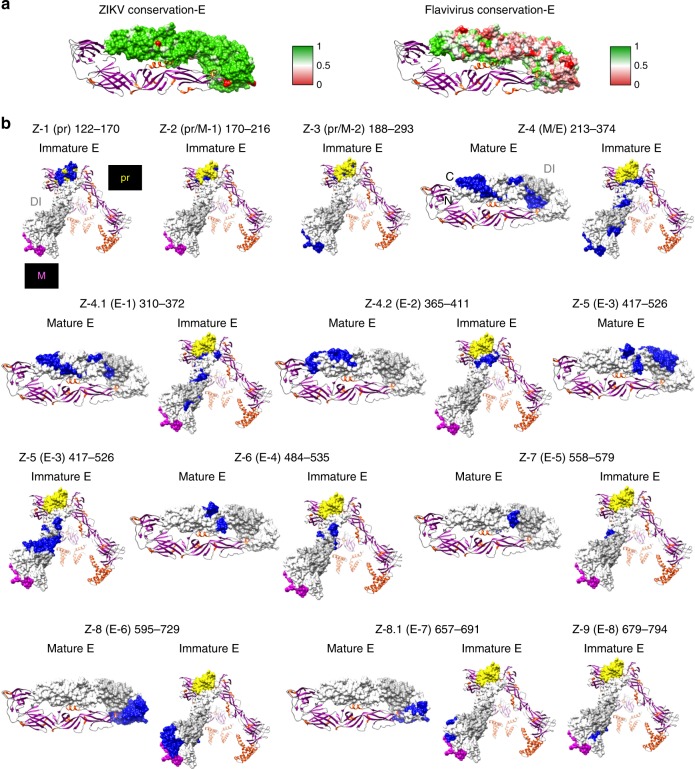
Fig. 3.
Structural representation of antigenic sites identified in ZIKV-E protein. a (Left panel) Heat map showing sequence conservation on one monomer chain of mature ZIKV-E protein structure (PDB# 5JHM) based on comparison with several Zika virus (ZIKV) isolates (Paraiba, Uganda-1947, Nigeria-1968, Senegal-2001, Micronesia-2007, and Brazil-2016 strains) and (right panel) ZIKV vs. other flaviviruses (Dengue virus (DENV) 1 to 4, West Nile virus, Yellow Fever virus and ZIKV). The heat map has been color coded from red (0) to green (1), where green denotes complete conservation. b Antigenic sites within ZIKV-prM/E identified by the GFPDL analysis are depicted in blue on the structures of both immature (PDB 5U4W) and mature ZIKV-E (PDB 5JHM). Domain I is shaded in light gray (PDB 5JHM), pr domain is shaded in yellow and M in pink (PDB 5U4W). PDB Structure# 5U4W encompasses residues 288–794 and PDB structure# 5JHM encompasses residues 313–699 based on ZIKV_ICD polyprotein sequence (Supplementary Fig. 1)