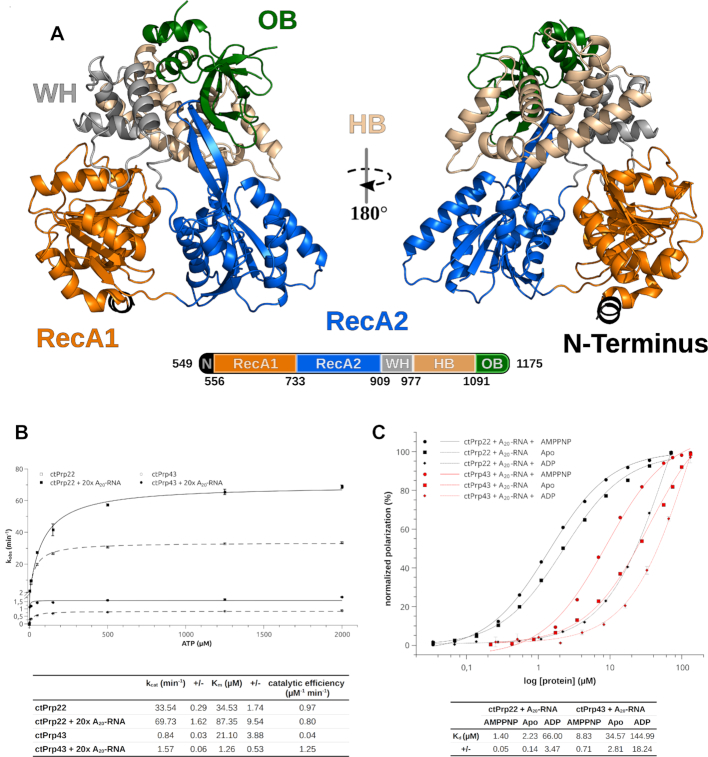
Figure 1.
Structural overview of Prp22 from Chaetomium thermophilum and functional comparison of ctPrp22 and ctPrp43. (A) Structural overview of Prp22 from C. thermophilum. The model of ctPrp22 is displayed as a cartoon representation. Residues from the truncated N-terminus (549–556) are shown in black, the RecA1 domain (557–733) in orange, the RecA2 domain (734–909) in blue, the WH domain (WH; 910–977) in gray, the HB domain (HB; 978–1091) in wheat and the OB fold (OB; 1092–1175) in green. (B) ATPase assays show that ctPrp22 has an increased ATPase activity compared to ctPrp43, but both are stimulated by the presence of an A20-RNA. (C) ctPrp22 and ctPrp43 show the same binding mode toward an A20-RNA in dependence of an adenosine nucleotide. Both bind the ssRNA best in presence of AMPPNP and show less binding in absence of any adenosine nucleotide. The affinity toward ssRNA is drastically decreased when ADP is present. ATPase activity as well as RNA-binding experiments were determined in triplicates and error bars for each measured data point are depicted. The error of fit is indicated in the tables as +/-.