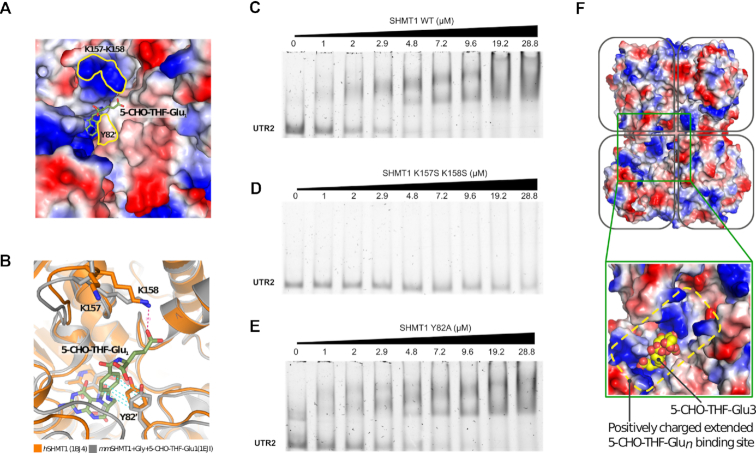
Figure 6.
SHMT1 amino acid residues involved in folate and RNA binding. (A) Surface representation of the folate-binding site of human SHMT1 (hSHMT, PDB id: 1BJ4 (53)) coloured by electrostatic potential (red are acidic and blue basic residues). The substrate (5-CHO-THF-Glu1) from mouse SHMT1 (mmSHMT1, PDB id: 1EJI (54)) is shown as sticks and superposed to the human enzyme. The location of K157, K158 and Y82′ is highlighted by the yellow contours. (B) Superposition of the human and mouse crystal structures (in orange and grey, respectively) showing the interaction of K158 and Y82′ with the folate substrate. (C–E) EMSA assays carried out by incubating 0.24 μM of unlabelled UTR2 with the indicated concentrations of SHMT1 wild-type (WT) (C), K157S-K158S (D) and Y82A (E) mutants. The apparent dissociation constants (Kdapp) of the protein–UTR2 complexes were measured for the WT and for the Y82A mutant, and are respectively 1.39 ± 0.30 μM and 3.0 ± 0.68 μM. The Kdapp value for the K157S-K158S SHMT1 mutant cannot be calculated given the low affinity of this protein for the UTR2 RNA in the concentration range explored. (F) Surface charge distribution of the human SHMT1 tetramer (PDB id: 1BJ4 (53)). In the blow up, 5-CHO-THF-Glu3, as observed in complex with rabbit SHMT1 (PDB id: 1LS3 (31)), is shown as yellow spheres superposed to the human enzyme. In the rabbit structure, the 5-CHO-THF-Glu3 is bound to two subunits (0.5 stoichiometry), with the poly-glutamylated tail making extensive and non-specific electrostatic interactions with the positively charged residues that surround the folate binding cleft.