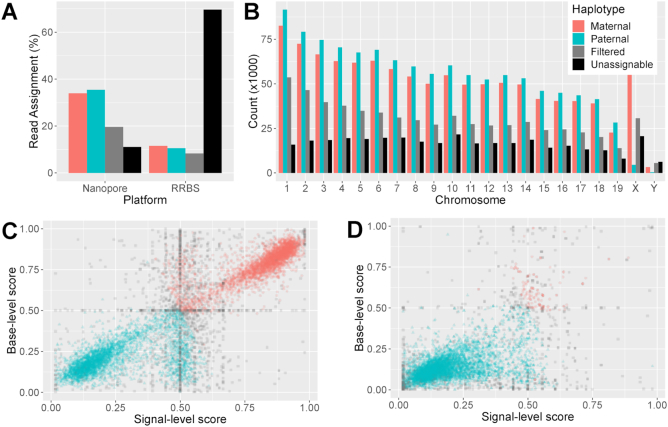
Figure 2.
Accurate and efficient haplotyping of nanopore reads. (A) Percentages of mapped reads from RRBS and nanopore sequencing that were assigned to the B6 genome (maternal), Cast genome (paternal), or that could not be haplotyped (filtered) for the B6 × Cast F1 sample. (B) Percentages of mapped reads from nanopore sequencing that were assigned to each haplotype on each chromosome. (C) Scatter plot of haplotype scores for nanopore reads according to signal (x-axis) and basecall (y-axis) methods. Only 10 000 randomly selected reads are shown for ease of visualization. (D) Signal and basecall haplotype scores for reads from the sequencing of the pure parental Cast strain.